Abstract
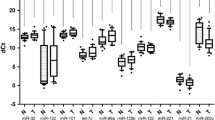
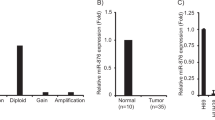
MicroRNAs (miRNAs) reportedly play significant roles in the progression of various cancers and hold huge potential as both diagnostic tools and therapeutic targets. Given the ongoing uncertainty surrounding the precise functions of several miRNAs in cholangiocarcinoma (CCA), this research undertakes a comprehensive analysis of CCA data sourced from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. The present study identified a novel miRNA, specifically miR-26b-3p, which exhibited prognostic value for individuals with CCA. Notably, miR-26b-3p was upregulated within CCA samples, with an inverse correlation established with patient prognosis (Hazard Ratio = 8.19, p = 0.018). Through a combination of functional enrichment analysis, analysis of the LncRNA-miR-26b-3p-mRNA interaction network, and validation by qRT PCR and western blotting, this study uncovered the potential of miR-26b-3p in potentiating the malignant progression of CCA via regulation of essential genes (including PSMD14, XAB2, SLC4A4) implicated in processes such as endoplasmic reticulum (ER) stress and responses to misfolded proteins. Our findings introduce novel and valuable insights that position miR-26b-3p-associated genes as promising biomarkers for the diagnosis and treatment of CCA.









Similar content being viewed by others
Data Availability
The data generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Razumilava, N., & Gores, G. J. (2014). Cholangiocarcinoma. Lancet, 383(9935), 2168–2179.
Xue, R., Chen, L., Zhang, C., Fujita, M., Li, R., Yan, S. M., et al. (2019). Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell, 35(6), 932–47.8.
Doherty, B., Nambudiri, V. E., & Palmer, W. C. (2017). Update on the diagnosis and treatment of cholangiocarcinoma. Current Gastroenterology Reports, 19(1), 2.
Tang, T. Y., Huang, X., Zhang, G., Lu, M. H., & Liang, T. B. (2022). mRNA vaccine development for cholangiocarcinoma: A precise pipeline. Military Medical Research, 9(1), 40.
Cillo, U., Fondevila, C., Donadon, M., Gringeri, E., Mocchegiani, F., Schlitt, H. J., et al. (2019). Surgery for cholangiocarcinoma. Liver International, 39(Suppl 1), 143–155.
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297.
Tafrihi, M., & Hasheminasab, E. (2019). MiRNAs: Biology, Biogenesis, their Web-based Tools, and Databases. MicroRNA, 8(1), 4–27.
Tutar, L., Ozgur, A., & Tutar, Y. (2018). Involvement of miRNAs and Pseudogenes in Cancer. Methods in Molecular Biology, 1699, 45–66.
Hussen, B. M., Rasul, M. F., Abdullah, S. R., Hidayat, H. J., Faraj, G. S. H., Ali, F. A., et al. (2023). Targeting miRNA by CRISPR/Cas in cancer: Advantages and challenges. Military Medical Research, 10(1), 32.
Li, Z., Shen, J., Chan, M. T., & Wu, W. K. (2017). The role of microRNAs in intrahepatic cholangiocarcinoma. Journal of Cellular and Molecular Medicine, 21(1), 177–184.
Cao, J., Sun, L., Li, J., Zhou, C., Cheng, L., Chen, K., et al. (2018). A novel three-miRNA signature predicts survival in cholangiocarcinoma based on RNA-Seq data. Oncology Reports, 40(3), 1422–1434.
Yao, Y., Jiao, D., Liu, Z., Chen, J., Zhou, X., Li, Z., et al. (2020). Novel miRNA Predicts Survival and Prognosis of Cholangiocarcinoma Based on RNA-seq Data and In Vitro Experiments. BioMed Research International, 2020, 5976127.
Wu, H. Y., Xia, S., Liu, A. G., Wei, M. D., Chen, Z. B., Li, Y. X., et al. (2019). Upregulation of miR-132-3p in cholangiocarcinoma tissues: A study based on RT-qPCR, The Cancer Genome Atlas miRNA sequencing, Gene Expression Omnibus microarray data and bioinformatics analyses. Molecular Medicine Reports, 20(6), 5002–5020.
Zhao, Y., Chen, J., Hao, Y., Wang, B., Wang, Y., Liu, Q., et al. (2022). Predicting the recurrence of chronic rhinosinusitis with nasal polyps using nasal microbiota. Allergy, 77(2), 540–549.
Liang, J. Y., Wang, D. S., Lin, H. C., Chen, X. X., Yang, H., Zheng, Y., et al. (2020). A novel ferroptosis-related gene signature for overall survival prediction in patients with Hepatocellular Carcinoma. International Journal of Biological Sciences, 16(13), 2430–2441.
Chen, D., Liu, J., Zang, L., Xiao, T., Zhang, X., Li, Z., et al. (2022). Integrated machine learning and bioinformatic analyses constructed a novel stemness-related classifier to predict prognosis and immunotherapy responses for hepatocellular carcinoma patients. International Journal of Biological Sciences, 18(1), 360–373.
Ranstam, J., & Cook, J. A. (2018). LASSO regression. British Journal of Surgery., 105, 1338–1348.
Muthukrishnan R, Rohini R. LASSO: A feature selection technique in predictive modeling for machine learning. 2016 Ieee International Conference on Advances in Computer Applications (Icaca). https://academic.oup.com/bjs/article/105/10/1348/6122951. https://doi.org/10.1002/bjs.10895
Kim, S. M., Kim, Y., Jeong, K., Jeong, H., & Kim, J. (2018). Logistic LASSO regression for the diagnosis of breast cancer using clinical demographic data and the BI-RADS lexicon for ultrasonography. Ultrasonography, 37(1), 36–42.
Vasaikar, S. V., Straub, P., Wang, J., & Zhang, B. (2018). LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Research, 46(D1), D956–D963.
Maragkakis, M., Vergoulis, T., Alexiou, P., Reczko, M., Plomaritou, K., Gousis, M., et al. (2011). DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Research, 39, 145–8.
Li, R., Deng, Y., Liang, J., Hu, Z., Li, X., Liu, H., et al. (2021). Circular RNA circ-102,166 acts as a sponge of miR-182 and miR-184 to suppress hepatocellular carcinoma proliferation and invasion. Cellular Oncology (Dordrecht), 44(2), 279–295.
Lin, Y., Jiang, M., Chen, W., Zhao, T., & Wei, Y. (2019). Cancer and ER stress: Mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomedicine Pharmacotherapy, 118, 109249.
Talabnin, K., Talabnin, C., Ishihara, M., & Azadi, P. (2018). Increased expression of the high-mannose M6N2 and NeuAc3H3N3M3N2F tri-antennary N-glycans in cholangiocarcinoma. Oncology Letters, 15(1), 1030–1036.
Sun, S. C. (2017). The non-canonical NF-kappaB pathway in immunity and inflammation. Nature Reviews Immunology, 17(9), 545–558.
Sirica, A. E. (2011). The role of cancer-associated myofibroblasts in intrahepatic cholangiocarcinoma. Nature Reviews Gastroenterology & Hepatology, 9(1), 44–54.
Zhang, L., Xu, H., Ma, C., Zhang, J., Zhao, Y., Yang, X., et al. (2020). Upregulation of deubiquitinase PSMD14 in lung adenocarcinoma (LUAD) and its prognostic significance. Journal of Cancer, 11(10), 2962–2971.
Li, Y., Huang, J., Zeng, B., Yang, D., Sun, J., Yin, X., et al. (2018). PSMD2 regulates breast cancer cell proliferation and cell cycle progression by modulating p21 and p27 proteasomal degradation. Cancer letters, 430, 109–122.
Zhang, Z., Li, H., Zhao, Y., Guo, Q., Yu, Y., Zhu, S., et al. (2019). Asporin promotes cell proliferation via interacting with PSMD2 in gastric cancer. Frontiers in Bioscience, 24, 1178–1189.
Fang, J., Rhyasen, G., Bolanos, L., Rasch, C., Varney, M., Wunderlich, M., et al. (2012). Cytotoxic effects of bortezomib in myelodysplastic syndrome/acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1. Blood, 120(4), 858–867.
Shitara, A., Shibui, T., Okayama, M., Arakawa, T., Mizoguchi, I., Sakakura, Y., et al. (2013). VAMP4 is required to maintain the ribbon structure of the Golgi apparatus. Molecular and Cellular Biochemistry, 380(1–2), 11–21.
Sun, J., Wang, L., Bao, H., Premi, S., Das, U., Chapman, E. R., et al. (2019). Functional cooperation of alpha-synuclein and VAMP2 in synaptic vesicle recycling. Proceedings of the National Academy of Sciences of the United States of America, 116(23), 11113–11115.
Ye, Y., Gu, B., Wang, Y., Shen, S., & Huang, W. (2019). E2F1-mediated MNX1-AS1-miR-218-5p-SEC61A1 feedback loop contributes to the progression of colon adenocarcinoma. Journal of Cellular Biochemistry, 120(4), 6145–6153.
Muller-McNicoll, M., Botti, V., de Jesus Domingues, A. M., Brandl, H., Schwich, O. D., Steiner, M. C., et al. (2016). SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes & Development, 30(5), 553–566.
Hu, W., Lei, L., Xie, X., Huang, L., Cui, Q., Dang, T., et al. (2019). Heterogeneous nuclear ribonucleoprotein L facilitates recruitment of 53BP1 and BRCA1 at the DNA break sites induced by oxaliplatin in colorectal cancer. Cell Death & Disease, 10(8), 550.
Song, X., Wan, X., Huang, T., Zeng, C., Sastry, N., Wu, B., et al. (2019). SRSF3-Regulated RNA Alternative Splicing Promotes Glioblastoma Tumorigenicity by Affecting Multiple Cellular Processes. Cancer Research, 79(20), 5288–5301.
Mano, Y., Aishima, S., Fukuhara, T., Tanaka, Y., Kubo, Y., Motomura, T., et al. (2013). Decreased roundabout 1 expression promotes development of intrahepatic cholangiocarcinoma. Human Pathology, 44(11), 2419–2426.
Liu, J., Liu, W., Li, H., Deng, Q., Yang, M., Li, X., et al. (2019). Identification of key genes and pathways associated with cholangiocarcinoma development based on weighted gene correlation network analysis. PeerJ, 7, e7968.
Takahashi, R. U., Prieto-Vila, M., Kohama, I., & Ochiya, T. (2019). Development of miRNA-based therapeutic approaches for cancer patients. Cancer science, 110(4), 1140–1147.
Rupaimoole, R., & Slack, F. J. (2017). MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nature Reviews Drug Discovery, 16(3), 203–222.
Han, Y., Meng, F., Venter, J., Wu, N., Wan, Y., Standeford, H., et al. (2016). miR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. Journal of Hepatology, 64(6), 1295–1304.
Lampis, A., Carotenuto, P., Vlachogiannis, G., Cascione, L., Hedayat, S., Burke, R., et al. (2018). MIR21 Drives Resistance to Heat Shock Protein 90 Inhibition in Cholangiocarcinoma. Gastroenterology, 154(4), 1066–79.e5.
Pungpapong, V., Zhang, M., & Zhang, D. (2020). Integrating biological knowledge into case-control analysis through iterated conditional modes/medians algorithm. Journal of Computational Biology : A Journal of Computational Molecular Cell Biology, 27(7), 1171–1179.
Khosla, R., Hemati, H., Rastogi, A., Ramakrishna, G., Sarin, S. K., & Trehanpati, N. (2019). miR-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in HCC. Liver international : Official Journal of the International Association for the Study of the Liver, 39(9), 1692–1703.
Geng, F., Lu, G. F., Ji, M. H., Kong, D. Y., Wang, S. Y., Tian, H., et al. (2019). MicroRNA-26b-3p/ANTXR1 signaling modulates proliferation, migration, and apoptosis of glioma. American Journal of Translational Research, 11(12), 7568–7578.
Xie, T., Pi, G., Yang, B., Ren, H., Yu, J., Ren, Q., et al. (2019). Long non-coding RNA 520 is a negative prognostic biomarker and exhibits pro-oncogenic function in nasopharyngeal carcinoma carcinogenesis through regulation of miR-26b-3p/USP39 axis. Gene, 707, 44–52.
Satoh, J., Kino, Y., & Niida, S. (2015). MicroRNA-Seq data analysis pipeline to identify blood biomarkers for alzheimer’s disease from public data. Biomarker insights, 10, 21–31.
Hong, H., Li, Y., & Su, B. (2017). Identification of circulating mir-125b as a potential biomarker of alzheimer’s disease in APP/PS1 transgenic mouse. Journal of Alzheimer’s Disease : JAD, 59(4), 1449–1458.
Anaparti, V., Smolik, I., Meng, X., Spicer, V., Mookherjee, N., & El-Gabalawy, H. (2017). Whole blood microRNA expression pattern differentiates patients with rheumatoid arthritis, their seropositive first-degree relatives, and healthy unrelated control subjects. Arthritis Research & Therapy, 19(1), 249.
Rizvi, S., & Gores, G. J. (2013). Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology, 145(6), 1215–1229.
Funding
This work was supported by the National Natural Science Foundation of China, 81802897 and 81901943; the Natural Science Foundation of Guangdong Province, 2021A1515011156, 2021A1515012493; the Operating Foundation of Guangdong Provincial Key Laboratory of Liver Disease Research, 2020B1212060019; the Guangzhou Basic and Applied Basic Research Foundation, 202102020237 and Guangzhou Basic and Applied Basic Research Project Co-funded by Municipal Schools (institutes), 2023A03J0727. Science and Technology Project of Guangzhou, SL2024A03J01216. Scientific Reserch Project of Dongguan Binhaiwan Central Hospital, 2021001.
Author information
Authors and Affiliations
Contributions
Conception and design: Rong Li, Xijing Yan, Kunpeng Hu, Xin Sui. Administrative support: Rong Li. Provision of study materials or patients: Zhongying Hu, Jinliang Liang, Xuejiao Li, Jiao Gong, Jun Zheng. Collection and assembly of data: Rong Li, Xijing Yan, Zhongying Hu, Jinliang Liang. Data analysis and interpretation: Rong Li, Xijing Yan, Kunpeng Hu, Xin Sui, Zhongying Hu, Jinliang Liang, Jiao Gong, Jun Zheng. Manuscript writing: All authors.
Corresponding authors
Ethics declarations
Ethical Statement
Our research was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University. All CCA data involved in this research were retrieved from the online databases (NCBI GEO DataSets and TCGA) which could be confirmed that all written informed consent had already been obtained and the data acquisition were followed the related data access policy and published guidelines.
Consent to Participate
The authors agreed to participate in this work.
Consent for Publication
The authors agreed to publish this work.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, X., Hu, Z., Li, X. et al. Systemic analysis of the prognostic significance and interaction network of miR-26b-3p in cholangiocarcinoma. Appl Biochem Biotechnol (2023). https://doi.org/10.1007/s12010-023-04753-x
Accepted:
Published:
DOI: https://doi.org/10.1007/s12010-023-04753-x