Abstract
Objective
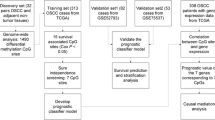
This study aimed to examine a novel method for prognostic evaluation of patients with oral squamous cell carcinoma (OSCC) based on the expression of heterogeneous nuclear ribonucleoprotein C (HNRNPC), YTH domain-binding protein 2 (YTHDF2), and methyltransferase 14 (METTL14).
Methods
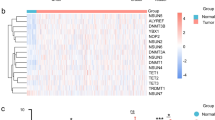
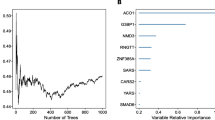
We obtained the RNA sequence and clinical information of OSCC patients from The Cancer Genome Atlas database. An optical method was established by the least absolute shrinkage and selection operator Cox regression algorithm, which was used to calculate the risk score of every sample. In addition, all samples (n=239) were classified into high-risk (n=119) and low-risk (n=120) groups, and the overall survival (OS) time and clinical characteristics were compared between groups. Moreover, bioinformatics analysis was carried out. Gene set enrichment analysis was performed to investigate the signaling pathways of HNRNPC, YTHDF2, and METTL14.
Results
The two groups showed significantly different OS time, tumor grades, tumor stages, and pathologic T stages (P<0.05). The receiver operating characteristic analysis identified that our method was effective and it was more accurate than use of age, gender, tumor grade, tumor stage, pathologic T stage, and pathologic N stage in OSCC prognostic prediction. Gene set enrichment analysis revealed that HNRNPC, YTHDF2, and METTL14 were mainly associated with ubiquitin-mediated proteolysis, cell cycle, RNA degradation, and spliceosome signaling pathways.
Conclusion
The method based on the expression of HNRNPC, YTHDF2, and METTL14 can predict the prognosis of patients with OSCC independently, and its prognostic value is better than that of clinicopathological characteristic indicators.
Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2018,68(6):394–424
Bloebaum M, Poort L, Böckmann R, et al. Survival after curative surgical treatment for primary oral squamous cell carcinoma. J Craniomaxillofac Surg, 2014,42(8):1572–1576
Fuller CD, Wang SJ, Thomas CR, et al. Conditional survival in head and neck squamous cell carcinoma: results from the SEER dataset 1973–1998. Cancer, 2007,109(7):1331–1343
Arduino PG, Carrozzo M, Chiecchio A, et al. Clinical and histopathologic independent prognostic factors in oral squamous cell carcinoma: A retrospective study of 334 cases. J Oral Maxillofac Surg, 2008,66(8):1570–1579
He C. Grand challenge commentary: RNA epigenetics. Nat Chem Biol, 2010,6(11):863–865
Ke S, Pandya-Jones A, Saito Y, et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev, 2017,31(10):990–1006
Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol, 2016,23(2):98–102
Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res, 2018,28(6):616–624
Cui Q, Shi H, Ye P, et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep, 2017,18(11):2622–2634
Chai RC, Wu F, Wang QX, et al. m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging, 2019,11(4):1204–1225
Pan Y, Ma P, Liu Y, et al. Multiple functions of m6A RNA methylation in cancer. J Hematol Oncol, 2018,11(1):48–58
Dai DJ, Wang HY, Zhu LY, et al. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis, 2018,9(2):124–136
Xu L, Yu C, Du XJ. Bioinformatics analysis of m6A methylation regulators’ influence on survival prognosis of patients with oral squamous cell carcinoma. Acta Med Univ Sci Technol Huazhong (Chinese), 2020,49(4):443–449
Zhao W, Cui Y, Liu L, et al. METTL3 facilitates oral squamous cell carcinoma Tumorigenesis by enhancing c-Myc stability via YTHDF1-mediated m6A modification. Mol Ther Nucleic Acids, 2020,20(6):1–12
Liu L, Wu Y, Li Q, et al. METTL3 Promotes Tumorigenesis and Metastasis through BMI1 m6A Methylation in Oral Squamous Cell Carcinoma. Mol Ther, 2020,28(10):2177–2190
Huang GZ, Wu QQ, Zheng ZN, et al. m6A-related bioinformatics analysis reveals that HNRNPC facilitates progression of OSCC via EMT. Aging, 2020,12(12):11667–11684
Moore SR, Johnson NW, Pierce AM, et al. The epidemiology of mouth cancer: a review of global incidence. Oral Dis, 2000,6(2):65–74
Chen JX, Sun YC, Xu X, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle, 2017,16(23):2259–2271
Zhong L, Liao D, Zhang M, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett, 2019,442:252–261
Paris J, Morgan M, Campos J, et al. Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell, 2019,25(1):137–148
Wu YS, Zhao WW, Liu Y, et al. Function of HNRNPC in breast cancer cells by controlling the dsRNA-induced interferon response. EMBO J, 2018,37(23):e99017
Weng HY, Huang HL, Wu HZ, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell, 2018,22(2):191–205
Ma JZ, Yang F, Zhou C, et al. METTL14 suppresses the metastatic potential of HCC by modulating m6A-dependent primary miRNA processing. Hepatology, 2017,65(2):529–543
Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer, 2006,6(5):369–381
Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature, 2004,432(11):316–323
Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer, 2005,5(12):773–785
David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev, 2010,24(21):2343–2364
Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol, 2017,18(9):655–670
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
Additional information
The present study was supported by the National Natural Science Foundation of China (No. 81802710).
Rights and permissions
About this article
Cite this article
Xu, L., Yu, C. & Du, Xj. Prognostic Evaluation for Oral Squamous Cell Carcinoma: A Novel Method Based on m6A Methylation Regulators. CURR MED SCI 42, 841–846 (2022). https://doi.org/10.1007/s11596-022-2611-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-022-2611-7