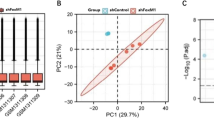
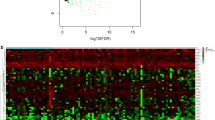
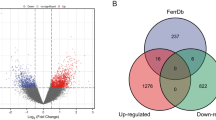
Abstract
Objective
Osteosarcoma is one of the most common types of bone sarcoma with a poor prognosis. However, identifying the predictive factors that contribute to the response to neoadjuvant chemotherapy remains a significant challenge.
Methods
A public data series (GSE87437) was downloaded to identify differentially expressed genes (DEGs) and differentially expressed lncRNAs (DElncRNAs) between osteosarcoma patients that do and do not respond to preoperative chemotherapy. Subsequently, functional analysis of the transcriptome expression profile, regulatory networks of DEGs and DElncRNAs, competing endogenous RNAs (ceRNA) and protein-protein interaction networks were performed. Furthermore, the function, pathway, and survival analysis of hub genes was performed and drug and disease relationship prediction of DElncRNA was carried out.
Results
A total of 626 DEGs, 26 DElncRNAs, and 18 hub genes were identified. However, only one gene and two lncRNAs were found to be suitable as candidate gene and lncRNAs respectively.
Conclusion
The DEGs, hub genes, candidate gene, and candidate lncRNAs screened out in this context were considered as potential biomarkers for the response to neoadjuvant chemotherapy of osteosarcoma.
Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin, 2020,70(1):7–30
Close AG, Dreyzin A, Miller KD, et al. Adolescent and young adult oncology-past, present, and future. CA Cancer J Clin, 2019,69(6):485–496
Crompton JG, Ogura K, Bernthal NM, et al. Local Control of Soft Tissue and Bone Sarcomas. J Clin Oncol, 2018,36(2):111–117
Li M, Jin X, Guo F, et al. Integrative analyses of key genes and regulatory elements in fluoride-affected osteosarcoma. J Cell Biochem, 2019,120(9):15397–15409
Li M, Jin X, Li H, et al. Comprehensive Analysis of Key Genes and Regulatory Elements in Osteosarcoma Affected by Bone Matrix Mineral With Prognostic Values. Front Genet, 2020,11:533
Li M, Jin X, Li H, et al. Key genes with prognostic values in suppression of osteosarcoma metastasis using comprehensive analysis. BMC Cancer, 2020,20(1):65
Vella S, Tavanti E, Hattinger CM, et al. Targeting CDKs with Roscovitine Increases Sensitivity to DNA Damaging Drugs of Human Osteosarcoma Cells. PLoS One, 2016,11(11):e0166233
Clough E, Barrett T, The Gene Expression Omnibus Database. Methods Mol Biol, 2016,1418:93–110
Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA, 2005,102(43):15545–15550
Ge SX, Son EW, and Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics, 2018, 19(1):534
Zhou G, Xia J. Using OmicsNet for Network Integration and 3D Visualization. Curr Protoc Bioinformatics, 2019,65(1):e69
Casper J, Zweig AS, Villarreal C, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res, 2018,46(D1):D762–d769
Hu H, Miao YR, Jia LH, et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res, 2019,47(D1):D33–d38
Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics, 2011,27(3):431–432
Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med, 2018,16(1):264
Ma L, Cao J, Liu L, et al. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res, 2019,47(D1):D128–d134
Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One, 2018,13(10):e0206239
Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res, 2017,45(D1):D362–d368
Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson, 2012,14(1):83
Liao Y, Wang J, Jaehnig EJ, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res, 2019,47(W1):W199–w205
Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer, 2014,14:970
Li Y, Li L, Wang Z, et al. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res, 2018,46(3):1113–1123
Carvalho-Silva D, Pierleoni A, Pignatelli M, et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res, 2019,47(D1):D1056–d1065
Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res, 2017,45(W1): W98–W102
Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res, 2018,46(D1):D1074–D1082
Miwa S, Takeuchi A, Shirai T, et al. Prognostic value of radiological response to chemotherapy in patients with osteosarcoma. PLoS One, 2013,8(7):e70015
Byun BH, Kong CB, Lim I, et al. Early response monitoring to neoadjuvant chemotherapy in osteosarcoma using sequential 18F-FDG PET/CT and MRI. Eur J Nucl Med Mol Imaging, 2014,41(8):1553–1562
Laux CJ, Berzaczy G, Weber M, et al. Tumour response of osteosarcoma to neoadjuvant chemotherapy evaluated by magnetic resonance imaging as prognostic factor for outcome. Int Orthop, 2015,39(1):97–104
Zhang H, Ge J, Hong H, et al. Genetic polymorphisms in ERCC1 and ERCC2 genes are associated with response to chemotherapy in osteosarcoma patients among Chinese population: a meta-analysis. World J Surg Oncol, 2017,15(1):75
Jung SY, Kwak JO, Kim HW, et al. Calcium sensing receptor forms complex with and is up-regulated by caveolin-1 in cultured human osteosarcoma (Saos-2) cells. Exp Mol Med, 2005,37(2):91–100
Bernardini G, Laschi M, Serchi T, et al. Proteomics and phosphoproteomics provide insights into the mechanism of action of a novel pyrazolo[3,4-d] pyrimidine Src inhibitor in human osteosarcoma. Mol Biosyst, 2014,10(6):1305–1312
Villanueva F, Araya H, Briceno P, et al. The cancer-related transcription factor RUNX2 modulates expression and secretion of the matricellular protein osteopontin in osteosarcoma cells to promote adhesion to endothelial pulmonary cells and lung metastasis. J Cell Physiol, 2019,234(8):13659–13679
Maurizi G, Verma N, Gadi A, et al. Sox2 is required for tumor development and cancer cell proliferation in osteosarcoma. Oncogene, 2018,37(33):4626–4632
Liu H, Chen Y, Zhou F, et al. Sox9 regulates hyperexpression of Wnt1 and Fzd1 in human osteosarcoma tissues and cells. Int J Clin Exp Pathol, 2014,7(8):4795–805
Mandela P, Yankova M, Conti LH, et al. Kalrn plays key roles within and outside of the nervous system. BMC Neurosci, 2012,13:136
Dang M, Wang Z, Zhang R, et al. KALRN Rare and Common Variants and Susceptibility to Ischemic Stroke in Chinese Han Population. Neuromolecular Med, 2015. 17(3): 241–50.
Liu HQ, Shu X, Ma Q, et al. Identifying specific miRNAs and associated mRNAs in CD44 and CD90 cancer stem cell subtypes in gastric cancer cell line SNU-5. Int J Clin Exp Pathol, 2020,13(6):1313–1323
Nath A, Lau EYT, Lee AM, et al. Discovering long noncoding RNA predictors of anticancer drug sensitivity beyond protein-coding genes. Proc Natl Acad Sci U S A, 2019,116(44):22 020–22 029
Vishnubalaji R, Shaath H, Elkord E, et al. Long noncoding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFβ and focal adhesion kinase (FAK) signaling. Cell Death Discov, 2019,5:109
Di Agostino S, Valenti F, Sacconi A, et al. Long Non-coding MIR205HG Depletes Hsa-miR-590-3p Leading to Unrestrained Proliferation in Head and Neck Squamous Cell Carcinoma. Theranostics, 2018,8(7):1850–1868
Wan J, Deng D, Wang X, et al. LINC00491 as a new molecular marker can promote the proliferation, migration and invasion of colon adenocarcinoma cells. Onco Targets Ther, 2019,12:6471–6480
Liu J, Yao Y, Hu Z, et al. Transcriptional profiling of long-intergenic noncoding RNAs in lung squamous cell carcinoma and its value in diagnosis and prognosis. Mol Genet Genomic Med, 2019,7(12):e994
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that there is no conflict of interest with any financial organization or corporation or individual that can inappropriately influence this work.
Additional information
The study was supported by the grant from the Research Foundation of Tongji Hospital (No. 2019B17).
Supplementary data
Rights and permissions
About this article
Cite this article
Li, M., Cheng, Wt., Li, H. et al. Comprehensive Analysis of Key mRNAs and lncRNAs in Osteosarcoma Response to Preoperative Chemotherapy with Prognostic Values. CURR MED SCI 41, 916–929 (2021). https://doi.org/10.1007/s11596-021-2430-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-021-2430-2