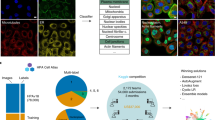
Abstract
Knowledge of protein expression in mammalian brains at regional and cellular levels can facilitate understanding of protein functions and associated diseases. As the mouse brain is a typical mammalian brain considering cell type and structure, several studies have been conducted to analyze protein expression in mouse brains. However, labeling protein expression using biotechnology is costly and time-consuming. Therefore, automated models that can accurately recognize protein expression are needed. Here, we constructed machine learning models to automatically annotate the protein expression intensity and cellular location in different mouse brain regions from immunofluorescence images. The brain regions and sub-regions were segmented through learning image features using an autoencoder and then performing K-means clustering and registration to align with the anatomical references. The protein expression intensities for those segmented structures were computed on the basis of the statistics of the image pixels, and patch-based weakly supervised methods and multi-instance learning were used to classify the cellular locations. Results demonstrated that the models achieved high accuracy in the expression intensity estimation, and the F1 score of the cellular location prediction was 74.5%. This work established an automated pipeline for analyzing mouse brain images and provided a foundation for further study of protein expression and functions.
Graphical Abstract







Similar content being viewed by others
References
Ding J, Ji J, Rabow Z, Shen T, Folz J, Brydges CR et al (2021) A metabolome atlas of the aging mouse brain. Nat Commun 12(1):6021. https://doi.org/10.1038/s41467-021-26310-y
Duan L, Liu J, Yin H, Wang W, Liu L, Shen J et al (2022) Dynamic changes in spatiotemporal transcriptome reveal maternal immune dysregulation of autism spectrum disorder. Comput Biol Med 151:106334. https://doi.org/10.1016/j.compbiomed.2022.106334
Sjöstedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C et al (2020) An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367(6482):eaay5947. https://doi.org/10.1126/science.aay5947
Zellner A, Müller SA, Lindner B, Beaufort N, Rozemuller AJ, Arzberger T et al (2022) Proteomic profiling in cerebral amyloid angiopathy reveals an overlap with CADASIL highlighting accumulation of HTRA1 and its substrates. Acta Neuropathol Commun 10(1):1–15
Ferreira M, Ventorim R, Almeida E, Silveira S, Silveira W (2021) Protein abundance prediction through machine learning methods. J Mol Biol 433(22):167267
Wang F, Wei L (2022) Multi-scale deep learning for the imbalanced multi-label protein subcellular localization prediction based on immunohistochemistry images. Bioinformatics 38(9):2602–2611
Cong H, Liu H, Chen Y, Cao Y (2020) Self-evoluting framework of deep convolutional neural network for multilocus protein subcellular localization. Med Biol Eng Compu 58:3017–3038
Xue Z-Z, Li C, Luo Z-M, Wang S-S, Xu Y-Y (2022) Automated classification of protein expression levels in immunohistochemistry images to improve the detection of cancer biomarkers. BMC Bioinformatics 23(1):470. https://doi.org/10.1186/s12859-022-05015-z
Ouyang W, Winsnes CF, Hjelmare M, Cesnik AJ, Åkesson L, Xu H et al (2019) Analysis of the human protein atlas image classification competition. Nat Methods 16(12):1254–1261. https://doi.org/10.1038/s41592-019-0658-6
Wang G, Xue M-Q, Shen H-B, Xu Y-Y (2022) Learning protein subcellular localization multi-view patterns from heterogeneous data of imaging, sequence and networks. Briefings in Bioinformatics 23(2):bbab539. https://doi.org/10.1093/bib/bbab539
Xue M-Q, Zhu X-L, Wang G, Xu Y-Y (2022) DULoc: quantitatively unmixing protein subcellular location patterns in immunofluorescence images based on deep learning features. Bioinformatics 38(3):827–833. https://doi.org/10.1093/bioinformatics/btab730
Giacopelli G, Migliore M, Tegolo D (2023) NeuronAlg: an innovative neuronal computational model for immunofluorescence image segmentation. Sensors 23(10):4598. https://doi.org/10.3390/s23104598
Goubran M, Leuze C, Hsueh B, Aswendt M, Ye L, Tian Q et al (2019) Multimodal image registration and connectivity analysis for integration of connectomic data from microscopy to MRI. Nat Commun 10(1):5504. https://doi.org/10.1038/s41467-019-13374-0
Li Y, Zhang Q, Zhou H, Li J, Li X, Li A (2023) Cerebrovascular segmentation from mesoscopic optical images using Swin Transformer. J Innov Opt Health Sci 2350009. https://doi.org/10.1142/S1793545823500098
Digre A, Lindskog C (2021) The human protein atlas—spatial localization of the human proteome in health and disease. Protein Sci 30(1):218–233
Tyson AL, Margrie TW (2022) Mesoscale microscopy and image analysis tools for understanding the brain. Prog Biophys Mol Biol 168:81–93. https://doi.org/10.1016/j.pbiomolbio.2021.06.013
Agarwal N, Xu X, Gopi M (2018) Geometry processing of conventionally produced mouse brain slice images. J Neurosci Methods 306:45–56. https://doi.org/10.1016/j.jneumeth.2018.04.008
Maintz JBA, Viergever MA (1998) A survey of medical image registration. Med Image Anal 2(1):1–36. https://doi.org/10.1016/S1361-8415(01)80026-8
Ni H, Tan C, Feng Z, Chen S, Zhang Z, Li W et al (2020) A robust image registration interface for large volume brain atlas. Sci Rep 10(1):2139. https://doi.org/10.1038/s41598-020-59042-y
Hirai R, Mori S, Suyari H, Tsuji H, Ishikawa H (2023) Optimizing 3DCT image registration for interfractional changes in carbon-ion prostate radiotherapy. Sci Rep 13(1):7448. https://doi.org/10.1038/s41598-023-34339-w
Chen Z, Zheng Y, Gee JC (2023) TransMatch: a transformer-based multilevel dual-stream feature matching network for unsupervised deformable image registration. IEEE Trans Med Imaging 1–1. https://doi.org/10.1109/TMI.2023.3288136
Toki MI, Cecchi F, Hembrough T, Syrigos KN, Rimm DL (2017) Proof of the quantitative potential of immunofluorescence by mass spectrometry. Lab Invest 97(3):329–334
Pécot T, Cuitiño MC, Johnson RH, Timmers C, Leone G (2022) Deep learning tools and modeling to estimate the temporal expression of cell cycle proteins from 2D still images. PLoS Comput Biol 18(3):e1009949
De León Rodríguez SG, Hernández Herrera P, Aguilar Flores C, Pérez Koldenkova V, Guerrero A, Mantilla A et al (2022) A machine learning workflow of multiplexed immunofluorescence images to interrogate activator and tolerogenic profiles of conventional type 1 dendritic cells infiltrating melanomas of disease-free and metastatic patients. J Oncol 2022:9775736
Fang K, Li C, Wang J (2023) An automatic immunofluorescence pattern classification framework for HEp-2 image based on supervised learning. Brief Bioinform 24(3):bbad144. https://doi.org/10.1093/bib/bbad144
Yang Y, Tu Y, Lei H, Long W (2023) HAMIL: hierarchical aggregation-based multi-instance learning for microscopy image classification. Pattern Recogn 136:109245. https://doi.org/10.1016/j.patcog.2022.109245
Abdi IY, Bartl M, Dakna M, Abdesselem H, Majbour N, Trenkwalder C et al (2023) Cross-sectional proteomic expression in Parkinson’s disease-related proteins in drug-naïve patients vs healthy controls with longitudinal clinical follow-up. Neurobiol Dis 177:105997. https://doi.org/10.1016/j.nbd.2023.105997
Uras I, Karayel-Basar M, Sahin B, Baykal AT (2023) Detection of early proteomic alterations in 5xFAD Alzheimer’s disease neonatal mouse model via MALDI-MSI. Alzheimers Dement 19(10):4572–4589
Taguchi K, Watanabe Y, Tsujimura A, Tanaka M (2019) Expression of alpha-synuclein is regulated in a neuronal cell type-dependent manner. Anat Sci Int 94(1):11–22. https://doi.org/10.1007/s12565-018-0464-8
Zahid S, Oellerich M, Asif AR, Ahmed N (2014) Differential expression of proteins in brain regions of Alzheimer’s Disease Patients. Neurochem Res 39(1):208–215. https://doi.org/10.1007/s11064-013-1210-1
Feng Y, Zhang L, Mo J (2020) Deep manifold preserving autoencoder for classifying breast cancer histopathological images. IEEE/ACM Trans Comput Biol Bioinform 17(1):91–101. https://doi.org/10.1109/TCBB.2018.2858763
Karim MR, Beyan O, Zappa A, Costa IG, Rebholz-Schuhmann D, Cochez M et al (2021) Deep learning-based clustering approaches for bioinformatics. Brief Bioinform 22(1):393–415. https://doi.org/10.1093/bib/bbz170
Wang C-W, Ka S-M, Chen A (2014) Robust image registration of biological microscopic images. Sci Rep 4(1):1–12
Agarwal N, Xu X, Gopi M (2016) Robust registration of mouse brain slices with severe histological artifacts. Proceedings of the Tenth Indian Conference on Computer Vision, Graphics and Image Processing p. 1–8
Kindle LM, Kakadiaris IA, Ju T, Carson JP (2011) A semiautomated approach for artefact removal in serial tissue cryosections. J Microsc 241(2):200–206
Besl PJ, McKay ND (1992) A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell 14(2):239–256. https://doi.org/10.1109/34.121791
Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N et al (2015) Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci 18(12):1819–1831
Wang D, Khosla A, Gargeya R, Irshad H, Beck AH (2016) Deep learning for identifying metastatic breast cancer. arXiv preprint arXiv:160605718
Frade J, Pereira T, Morgado J, Silva F, Freitas C, Mendes J et al (2022) Multiple instance learning for lung pathophysiological findings detection using CT scans. Med Biol Eng Compu 60(6):1569–1584. https://doi.org/10.1007/s11517-022-02526-y
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556
Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH (2016) Patch-based convolutional neural network for whole slide tissue image classification. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) p. 2424–2433
Wang X, Chen H, Gan C, Lin H, Dou Q, Tsougenis E et al (2020) Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans Cybern 50(9):3950–3962. https://doi.org/10.1109/TCYB.2019.2935141
Ilse M, Tomczak J, Welling M (2018) Attention-based deep multiple instance learning. International conference on machine learning: PMLR p. 2127–2136
Su Z, Tavolara TE, Carreno-Galeano G, Lee SJ, Gurcan MN, Niazi MKK (2022) Attention2majority: weak multiple instance learning for regenerative kidney grading on whole slide images. Med Image Anal 79:102462. https://doi.org/10.1016/j.media.2022.102462
Yao J, Zhu X, Jonnagaddala J, Hawkins N, Huang J (2020) Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med Image Anal 65:101789. https://doi.org/10.1016/j.media.2020.101789
Lin TY, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. 2017 IEEE International Conference on Computer Vision (ICCV) p. 2999–3007
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) p. 770–778
Huang G, Liu Z, Maaten LVD, Weinberger KQ (2017) Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) p. 2261–2269
Tan M, Le Q (2019) Efficientnet: rethinking model scaling for convolutional neural networks. International conference on machine learning: PMLR p. 6105–6114
Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P (2017) Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph 61:2–13
Xu Y-Y, Yang F, Zhang Y, Shen H-B (2015) Bioimaging-based detection of mislocalized proteins in human cancers by semi-supervised learning. Bioinformatics 31(7):1111–1119
Pluim JP, Maintz JB, Viergever MA (2003) Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging 22(8):986–1004. https://doi.org/10.1109/TMI.2003.815867
Krstinic D, Braović M, Šerić L, Božić-Štulić D (2020) Multi-label classifier performance evaluation with confusion matrix. Comput Sci Inform Technol 10:1
Zhu X-L, Bao L-X, Xue M-Q, Xu Y-Y (2023) Automatic recognition of protein subcellular location patterns in single cells from immunofluorescence images based on deep learning. Brief Bioinform 24(1):bbac609. https://doi.org/10.1093/bib/bbac609
Stadler C, Rexhepaj E, Singan VR, Murphy RF, Pepperkok R, Uhlén M et al (2013) Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods 10(4):315–323. https://doi.org/10.1038/nmeth.2377
Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H et al (2017) A subcellular map of the human proteome. Science 356(6340):eaal3321
Tu Y, Lei H, Shen H-B, Yang Y (2022) SIFLoc: a self-supervised pre-training method for enhancing the recognition of protein subcellular localization in immunofluorescence microscopic images. Briefings in Bioinformatics 23(2):bbab605. https://doi.org/10.1093/bib/bbab605
Long W, Yang Y, Shen H-B (2020) ImPLoc: a multi-instance deep learning model for the prediction of protein subcellular localization based on immunohistochemistry images. Bioinformatics 36(7):2244–2250. https://doi.org/10.1093/bioinformatics/btz909
Nanni L, Paci M, Brahnam S, Lumini A (2022) Feature transforms for image data augmentation. Neural Comput Appl 34(24):22345–22356. https://doi.org/10.1007/s00521-022-07645-z
Nanni L, Paci M, Brahnam S, Lumini A (2021) Comparison of different image data augmentation approaches. J Imaging 7(12):254
Funding
This work was supported in part by the Natural Science Foundation of Guangdong Province of China (grant number 2022A1515011436) and the Guangzhou Municipal Science and Technology Project (grant number 202102021087).
Author information
Authors and Affiliations
Contributions
Y.Y. Xu designed the research. L.X. Bao and X.L. Zhu wrote the source code and performed the experiments. L.X. Bao, Z.M. Luo, and Y.Y. Xu wrote, edited, and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bao, LX., Luo, ZM., Zhu, XL. et al. Automated identification of protein expression intensity and classification of protein cellular locations in mouse brain regions from immunofluorescence images. Med Biol Eng Comput 62, 1105–1119 (2024). https://doi.org/10.1007/s11517-023-02985-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02985-x