Abstract
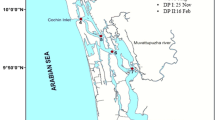
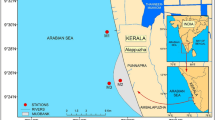
Mud banks and upwelling are two important oceanographic features occurring along the southwest coast of India during the southwest monsoon period. The study region, Alappuzha lying on the southwest coast of India, is unique due to the co-existence of upwelling and mud banks during the monsoon (MON) season. Water samples were collected from three stations, M1, M2, and M3, from April to September 2014, at weekly/biweekly intervals to determine the total bacterial abundance, viable prokaryotic counts, and total plate counts, along with measurements on physico-chemical parameters. For determining the heterotrophic culturable bacterial diversity, water samples were collected during two seasons, monsoon and pre-monsoon (PRM), from three stations. Water samples were inoculated into two non-selective broths for enrichment, DNA was extracted, and next-generation sequencing analysis was performed using Illumina Miseq sequencing. The sequence analysis revealed that dominant communities were Proteobacteria, followed by Firmicutes and Fusobacteria. Proportions of Fusobacteria increased during monsoon and proportions of Firmicutes were high in premonsoon season. Among Proteobacteria, Gammaproteobacteri is presented more than 99% of all the classes, irrespective of seasons. Vibrio was the most dominant genus during both seasons. The presence of anaerobic genera such as Propionigenium and Cetobacterium at all the stations during MON indicated the presence of upwelled waters. The genus Stenotrophomonas was observed in the M2 station alone. This study provides an overview of the culturable heterotrophic bacterial communities in a region in the southeastern Arabian Sea with coexisting mud banks and upwelling. The results of this study were compared with a published report on culture-independent bacterial diversity (from environmental DNA) from the same region. The study demonstrates that the use of culture media underrepresented the phylogenetic diversity and selectively enriched the class Gammaproteobacteria alone.







Similar content being viewed by others
References
Alfonso-Gordillo G, Cristiani-Urbina E, Flores-Ortiz CM, Peralta H, Cancino-Díaz JC, Cruz-Maya JA, Jan-Roblero J (2016) Stenotrophomonas maltophilia isolated from gasoline-contaminated soil is capable of degrading methyl tert-butyl ether. Electron J Biotechnol 23:12–20. https://doi.org/10.1016/j.ejbt.2016.06.006
Alonso-Sáez L, Gasol JM, Arístegui J, Vilas JC, Vaqué D, Duarte CM, Agustí S (2007) Large-scale variability in surface bacterial carbon demand and growth efficiency in the subtropical northeast Atlantic Ocean. Limnol Oceanogr. 52(2):533–546. https://doi.org/10.4319/lo.2007.52.2.0533
Alvarez-Yela AC, Mosquera-Rendón J, Noreña-P A, Cristancho M, López-Alvarez D (2019) Microbial diversity exploration of marine hosts at Serrana Bank, a coral atoll of the Seaflower Biosphere Reserve. Front Mar Sci. 6:338. https://doi.org/10.3389/fmars.2019.00338
Anas A, Chekidhenkuzhiyil J, Nilayangod C, Krishna K, Raju GT (2021) Prevalence of obligate and facultative anaerobic bacteria in the mudbank along the southwest coast of India. Regional Studies in Marine Science 42:101660. https://doi.org/10.1016/j.rsma.2021.101660
Anthony EJ, Dolique F, Gardel A, Gratiot N, Proisy C, Polidori L (2008) Nearshore intertidal topography and topographic-forcing mechanisms of an Amazon-derived Mud bank in French Guiana. Cont Shelf Res 28(6):813–822. https://doi.org/10.1016/j.csr.2008.01.003
Balachandran KK (2004) Does subterranean flow initiate mud banks off the southwest coast of India? Estuar Coast Shelf Sci 59(4):589–598. https://doi.org/10.1016/j.ecss.2003.11.004
Bano N, Hollibaugh JT (2002) Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl Environ Microbiol 68(2):505–518. https://doi.org/10.1128/AEM.68.2.505-518.2002
Benlloch S, Martínez-Murcia AJ, Rodríguez-Valera F (1995) Sequencing of bacterial and archaeal 16S rRNA genes directly amplified from a hypersaline environment. Syst Appl Microbiol 18(4):574–581. https://doi.org/10.1016/S0723-2020(11)80418-2
Benner R, Biddanda B (1998) Photochemical transformations of surface and deep marine dissolved organic matter: effects on bacterial growth. Limnol Oceanogr 43(6):1373–1378. https://doi.org/10.4319/lo.1998.43.6.1373
Bergen B, Herlemann DP, Jürgens K (2015) Zonation of bacterioplankton communities along aging upwelled water in the northern Benguela upwelling. Front Microbiol 6:621. https://doi.org/10.3389/fmicb.2015.00621
Bertagnolli AD, Treusch AH, Mason OU, Stingl U, Vergin KL, Chan F, Beszteri B, Giovannoni SJ (2011) Bacterial diversity in the bottom boundary layer of the inner continental shelf of Oregon, USA. Aqua Microb Ecol 64(1):15–25. https://doi.org/10.3354/ame01504
Bolhuis H, Stal LJ (2011) Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing. ISME J 5(11):1701–1712. https://doi.org/10.1038/ismej.2011.52
Burkill PH, Mantoura RFC, Owens NJP (1993) Biogeochemical cycling in the northwestern Indian Ocean: a brief overview. Deep Sea Res Part II: Topic. Stud Oceanogr 40(3):643–649. https://doi.org/10.1016/0967-0645(93)90049-S
Campbell BJ, Kirchman DL (2013) Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7(1):210–220. https://doi.org/10.1038/ismej.2012.93
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Clarke KR, Warwick RM (2001) A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Marine ecology Progress series 216:265–278. https://doi.org/10.3354/meps216265
Connon SA, Giovannoni SJ (2002) High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68(8):3878–3885. https://doi.org/10.1128/AEM.68.8.3878-3885.2002
Cragg BA, Parkes RJ (2014) Bacterial and Archaeal direct counts: a faster method of enumeration, for enrichment cultures and aqueous environmental samples. J Microbiol Meth 98:35–40. https://doi.org/10.1016/j.mimet.2013.12.006
Cury JC, Araujo FV, Coelho-Souza SA, Peixoto RS, Oliveira JA, Santos HF et al (2011) Microbial diversity of a Brazilian coastal region influenced by an upwelling system and anthropogenic activity. PLoS One 6(1):e16553. https://doi.org/10.1371/journal.pone.0016553
Dayana M, Gireeshkumar TR, Udayakrishnan PB, Muraleedharan KR, Madhu NV, Nair M et al (2019) Inter-annual variations in the hydrochemistry of Alappuzha mud banks, southwest coast of India. Cont Shelf Res 177:42–49. https://doi.org/10.1016/j.csr.2019.03.008
Diaz RJ, Rosenberg R (2008) Spreading dead zones and consequences for marine ecosystems. Science 321(5891):926–929. https://doi.org/10.1126/science.1156401
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345–366. https://doi.org/10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2
Eilers H, Pernthaler J, Glöckner FO, Amann R (2000) Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbial 66(7):3044–3051. https://doi.org/10.1128/AEM.66.7.3044-3051.2000
Ferguson RL, Buckley EN, Palumbo AV (1984) Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbial 47(1):49–55
Fernandes GL, Shenoy BD, Damare SR (2020) Diversity of bacterial community in the oxygen minimum zones of Arabian Sea and Bay of Bengal as deduced by illumina sequencing. Frontiers in microbiology 10:3153. https://doi.org/10.3389/fmicb.2019.03153
Fuchs BM, Woebken D, Zubkov MV, Burkill P, Amann R (2005) Molecular identification of picoplankton populationsin contrasting waters of the Arabian Sea. Aqua Microb Ecol 39(2):145–157. https://doi.org/10.3354/ame039145
Gerson VJ, Madhu NV, Jyothibabu R, Balachandran KK, Nair M, Revichandran C (2014) Oscillating environmental responses of the eastern Arabian Sea. Indian Journal of Geo-Marine Science Vol. 43. http://nopr.niscair.res.in/handle/123456789/26428
Gireeshkumar TR, Mathew D, Pratihary AK, Naik H, Narvekar KU, Araujo J et al (2017) Influence of upwelling induced near shore hypoxia on the Alappuzha Mud banks, South West Coast of India. Cont Shelf Res 139:1–8. https://doi.org/10.1016/j.csr.2017.03.009
Glöckner I, Siebert C, Polerecky L, Munwes YY, Lott C, Häusler S et al (2012) Microbial and chemical characterization of underwater fresh water springs in the Dead Sea. PLoS One 7(6):e38319. https://doi.org/10.1371/journal.pone.0038319
Gopinathan CK, Qasim SZ (1974) Mud banks of Kerala-their formation and characteristics. Ind J Mar Sci 3(2):105–114 http://eprints.cmfri.org.in/id/eprint/7309
Grasshoff K (1999) Determination of nitrite, nitrate, oxygen, thiosulphate. In: Grasshoff K, Ehrhardt M, Kremling K (eds) Methods of seawater analysis. VerlagChemieWeinheim, NewYork, pp 139–142, 143–150, 61–72, 81–139–142, 143–150, 61–72, 84
Greer CW (2010) Bacterial diversity in hydrocarbon-polluted rivers, estuaries and sediments. In: Handbook of hydrocarbon and lipid microbiology
Grosche A, Sekaran H, Pérez-Rodríguez I, Starovoytov V, Vetriani C (2015) Cetiapacifica gen. nov., sp. nov., a chemolithoautotrophic, thermophilic, nitrate-ammonifying bacterium from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 65(4):1144–1150. https://doi.org/10.1099/ijs.0.000070
Guerrero-Feijóo E, Nieto-Cid M, Sintes E, Dobal-Amador V, Hernando-Morales V, Álvarez M, Balagué V, Varela MM (2017) Optical properties of dissolved organic matter relate to different depth-specific patterns of archaeal and bacterial community structure in the North Atlantic Ocean. FEMS Microbiol Ecol 93(1):fiw224. https://doi.org/10.1093/femsec/fiw224
Gutierrez T, Berry D, Teske A, Aitken MD (2016) Enrichment of Fusobacteria in sea surface oil slicks from the Deepwater Horizon oil spill. Microorganisms 4(3):24. https://doi.org/10.3390/microorganisms4030024
He K, Zhang X, Ren S, Sun J (2016) Identity mappings in deep residual networks. In: European conference on computer vision. Springer, Cham, pp 630–645
Jasna V, Nathan VK, Parvathi A (2020) Comparison of bacterial community structure in coastal and offshore waters of the Bay of Bengal, India. Reg Stud Mar Sci 39:101414. https://doi.org/10.1016/j.rsma.2020.101414
Joux F, Lebaron P (1997) Ecological implications of an improved direct viable count method for aquatic bacteria. Appl Environ Microbiol 63(9):3643–3647 PMID: 16535694
Jyothibabu R, Balachandran KK, Jagadeesan L, Karnan C, Arunpandi N, Naqvi SWA, Pandiyarajan RS (2018) Mud Banks along the southwest coast of India are not too muddy for plankton. Sci Rep 8(1):2544. https://doi.org/10.1038/s41598-018-20667-9
Kelly KM, Chistoserdov AY (2001) Phylogenetic analysis of the succession of bacterial communities in the Great South Bay (Long Island). FEMS Microbiol Ecol 35(1):85–95. https://doi.org/10.1111/j.1574-6941.2001.tb00791.x
Kumar R, Mishra A, Jha B (2019) Bacterial community structure and functional diversity in subsurface seawater from the western coastal ecosystem of the Arabian Sea, India. Gene 701:55–64. https://doi.org/10.1016/j.gene.2019.02.099
Kurup PG (1977) Studies on the physical aspects of the mudbanks along the Kerala coast with special reference to the Purakadmudbank. Bull Dep Mar Sci Univ Cochin 8(81):72
Labrenz M, Jost G, Jürgens K (2007) Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic–anoxic interface. Aquat Microb Ecol 46(2):177–190. https://doi.org/10.3354/ame046177
Lamont T, Barlow RG, Kyewalyanga MS (2014) Physical drivers of phytoplankton production in the southern Benguela upwelling system. Deep Sea Res Part I:Oceanogr Res Pap 90:1–16. https://doi.org/10.1016/j.dsr.2014.03.003
Liu JP, Li AC, Xu KH, Velozzi DM, Yang ZS, Milliman JD, DeMaster DJ (2006) Sedimentary features of the Yangtze River-derived along-shelf clinoform deposit in the East China Sea. Cont Shelf Res 26(17-18):2141–2156. https://doi.org/10.1016/j.csr.2006.07.013
Mallik TK, Mukherji KK, Ramachandran KK (1988) Sedimentology of the Kerala mud banks (fluid muds?). Mar Geol 80(1-2):99–118. https://doi.org/10.1016/0025-3227(88)90074-6
McBride RA, Taylor MJ, Byrnes MR (2007) Coastal morphodynamics and Chenier-Plain evolution in southwestern Louisiana, USA: a geomorphic model. Geomorphology 88(3-4):367–422. https://doi.org/10.1016/j.geomorph.2006.11.013
Minirani S, Kurup PG (2007) Energy attenuation of sea surface waves through generation of interface waves on viscoelastic bottom as in the mud banks, SW coast of India. J Indian Geophys Union 11:143–146
Moopantakath J, Imchen M, Siddhardha B, Kumavath R (2020) 16s rRNA metagenomic analysis reveals predominance of Crtl and CruF genes in Arabian Sea coast of India. Science of The Total Environment 743:140699. https://doi.org/10.1016/j.scitotenv.2020.140699.
Muraleedharan KR, Kumar PD, Kumar SP, Srijith B, John S, Kumar KN (2017) Observed salinity changes in the Alappuzha mud bank, southwest coast of India and its implication to hypothesis of Mud bank formation. Cont Shelf Res 137:39–45. https://doi.org/10.1016/j.csr.2017.01.015
Muraleedharan KR, Kumar PD, Kumar SP, John S, Srijith B, Kumar KA et al (2018) Formation mechanism of mud bank along the Southwest Coast of India. Estur Coasts 41(4):1021–1035. https://doi.org/10.1007/s12237-017-0340-0
Murty AVS, Rao DS, Regunathan A, Gopinathan CP, Mathew KJ (1984) Hypotheses on the mudbanks. CMFRI Bull 31:8–17
Nogales B, Lanfranconi MP, Piña-Villalonga JM, Bosch R (2011) Anthropogenic perturbations in marine microbial communities. FEMS Microbiol Rev 35(2):275–298. https://doi.org/10.1111/j.1574-6976.2010.00248.x
Olsen I (2014) The family Fusobacteriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes. Springer, Berlin. https://doi.org/10.1007/978-3-642-30120-9_213
Ozdal M, Ozdal OG, Algur OF, Kurbanoglu EB (2017) Biodegradation of α-endosulfan via hydrolysis pathway by Stenotrophomonasmaltophilia OG2. 3. Biotech. 7(2):1–7. https://doi.org/10.1007/s13205-017-0765-y
Paingankar MS, Ahire K, Mishra P, Rajpathak S, Deobagkar DD (2019) Microbial diversity of the Arabian Sea in the Oxygen minimum zones by metagenomics approach. bioRxiv 731828. https://doi.org/10.1101/731828
Parsons, T. R. (1984). M. Takahashi, and B. Hargrave: Biological Oceanographic Processes. Elsevier
Parvathi A, Jasna V, Aswathy VK, Nathan VK, Aparna S, Balachandran KK (2019) Microbial diversity in a coastal environment with co-existing upwelling and mud-banks along the south west coast of India. Mol Biol Rep 46(3):3113–3127. https://doi.org/10.1007/s11033-019-04766-y
Piggot AM, Klaus JS, Swart PK (2012) Microbial and Geochemical Characterization of Carbonate Mudbanks from Florida Bay. AAPG Search and Discovery Article #90153©2012 AAPG Hedberg Conference Microbial Carbonate Reservoir Characterization, Houston, Texas, 3-8 June 2012.
Qaisrani MM, Zaheer A, Mirza MS, Naqqash T, Qaisrani TB, Hanif MK, Rasool G, Malik KA, Ullah S, Jamal MS, Mirza Z (2019) A comparative study of bacterial diversity based on culturable and culture-independent techniques in the rhizosphere of maize (Zea mays L.). Saudi J Biol Sci 26(7):1344–1351. https://doi.org/10.1016/j.sjbs.2019.03.010
Raes EJ, Bodrossy L, Van De Kamp J, Bissett A, Ostrowski M, Brown MV et al (2018) Oceanographic boundaries constrain microbial diversity gradients in the South Pacific Ocean. Proc Nat Acad Sci 115(35):E8266–E8275. https://doi.org/10.1073/pnas.1719335115
Rajpathak SN, Banerjee R, Mishra PG, Khedkar AM, Patil YM, Joshi SR, Deobagkar DD (2018) An exploration of microbial and associated functional diversity in the OMZ and non-OMZ areas in the Bay of Bengal. J Biosci 43(4):635–648. https://doi.org/10.1007/s12038-018-9781-2
Rehnstam AS, Bäckman S, Smith DC, Azam F, Hagström Å (1993) Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol Ecol 11(3-4):161–166. https://doi.org/10.1111/j.1574-6968.1993.tb05806.x
Robarts RD, Carr GM (2009) Bacteria, Bacterioplankton.A2 - likens, gene E. In: Likens GE (ed) Encyclopedia of Inland Waters. Academic Press, Oxford, pp 193–200. https://doi.org/10.1016/B978-012370626-3.00124-1
Rosselló-Mora R, Thamdrup B, Schäfer H, Weller R, Amann R (1999) The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Systemat Appl Microbiol 22(2):237–248. https://doi.org/10.1016/S0723-2020(99)80071-X
Ryther JH (1969) Photosynthesis and fish production in the sea. Science 166(3901):72–76
Sachithanandam V, Saravanane N, Chandrasekar K, Karthick P, Lalitha P, Elangovan SS, Sudhakar M (2020) Microbial diversity from the continental shelf regions of the Eastern Arabian Sea: a metagenomic approach. Saudi J Biol Sci 27(8):2065–2075. https://doi.org/10.1016/j.sjbs.2020.06.011
Schä H, Bernard L, Courties C, Lebaron P, Servais P, Pukall R et al (2001) Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol Ecol 34(3):243–253. https://doi.org/10.1111/j.1574-6941.2001.tb00775.x
Schink B (2006) The Genus Propionigenium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, Schink B (eds) The Prokaryotes. Springer, 3951, New York, p 3948. https://doi.org/10.1007/978-1-4757-2191-1_57
Schut F, de Vries EJ, Gottschal JC, Robertson BR, Harder W, Prins RA, Button DK (1993) Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl.Environ.Microbial 59(7):2150–2160
Shankar D, Shetye SR (1997) On the dynamics of the Lakshadweep high and low in the southeastern Arabian Sea. J Geophy Res Oceans 102(C6):12551–12562. https://doi.org/10.1029/97JC00465
Silas EG (1984) Mudbanks of Kerala-Karnataka; need for integrated study. CMFRI Bull 31:2–7
Smitha BR, Sanjeevan VN, Vimalkumar KG, Revichandran C (2008) On the upwelling off the southern tip and along the west coast of India. J Coast Res 4(24):95–102. https://doi.org/10.2112/06-0779.1
Somboonna N, Wilantho A, Rerngsamran P, Tongsima S (2019) Marine bacterial diversity in coastal Sichang island, the upper Gulf of Thailand, in 2011 wet season. Front Mar Sci 6:308. https://doi.org/10.3389/fmars.2019.00308
Stevens H, Ulloa O (2008) Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbial 10(5):1244–1259. https://doi.org/10.1111/j.1462-2920.2007.01539.x
Tamura T, Horaguchi K, Saito Y, Nguyen VL, Tateishi M, Ta TKO, Nanayama F, Watanabe K (2010) Monsoon-influenced variations in morphology and sediment of a mesotidal beach on the Mekong River delta coast. Geomorphology 116(1-2):11–23. https://doi.org/10.1016/j.geomorph.2009.10.003
Teira E, Gasol JM, Aranguren-Gassis M, Fernández A, González J, Lekunberri I, Álvarez-Salgado XA (2008) Linkages between bacterioplankton community composition, heterotrophic carbon cycling and environmental conditions in a highly dynamic coastal ecosystem. Environ Microbial 10(4):906–917. https://doi.org/10.1111/j.1462-2920.2007.01509.x
Thompson FL, Iida T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol Rev 68(3):403–431. https://doi.org/10.1128/MMBR.68.3.403-431.2004
Tytgat B, Verleyen E, Obbels D, Peeters K, De Wever A, D’hondt S, De Meyer T, Van Criekinge W, Vyverman W, Willems A (2014) Bacterial diversity assessment in Antarctic terrestrial and aquatic microbial mats: a comparison between bidirectional pyrosequencing and cultivation. PLoS One 9(6):e97564. https://doi.org/10.1371/journal.pone.0097564
Urakawa H, Yoshida T, Nishimura M, Ohwada K (2000) Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ Microbiol 2(5):542–554. https://doi.org/10.1046/j.1462-2920.2000.00137.x
Vartoukian SR (2016) Cultivation strategies for growth of uncultivated bacteria. J Oral Biosci 58(4):143–149. https://doi.org/10.1016/j.job.2016.08.001
Vetriani C, Tran HV, Kerkhof LJ (2003) Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl Environ Microbiol 69(11):6481–6488. https://doi.org/10.1128/AEM.69.11.6481-6488.2003
Walsh JP, Nittrouer CA (2004) Mangrove-bank sedimentation in a mesotidal environment with large sediment supply, Gulf of Papua. Mar Geol 208(2-4):225–248. https://doi.org/10.1016/j.margeo.2004.04.010
Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, Tortell PD, Hallam SJ (2009) Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326(5952):578–582. https://doi.org/10.1126/science.1175309
Weidner S, Arnold W, Puhler A (1996) Diversity of uncultured microorganisms associated with the seagrass Halophilastipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol 62(3):766–771
Wiebinga CJ, Veldhuis MJ, De Baar HJ (1997) Abundance and productivity of bacterioplankton in relation to seasonal upwelling in the northwest Indian Ocean. Deep Sea Res Part I: Oceanogr Res Pap 44(3):451–476. https://doi.org/10.1016/S0967-0637(96)00115-X
Zhang X, Lin H, Wlang X, Austin B (2018) Significance of Vibrio species in the marine organic carbon cycle—a review. Sci China Earth Sci 61(10):1357–1368. https://doi.org/10.1007/s11430-017-9229-x
Zhou J, Richlen ML, Sehein TR, Kulis DM, Anderson DM, Cai Z (2018) Microbial community structure and associations during a marine dinoflagellate bloom. Front.Microbial. 9:1201. https://doi.org/10.3389/fmicb.2018.01201
Zubkov MV, Fuchs BM, Archer SD, Kiene RP, Amann R, Burkill PH (2001) Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ Microbiol 3(5):304–311. https://doi.org/10.1046/j.1462-2920.2001.00196.x
Acknowledgements
The authors are grateful to the Director, NIO, Goa, and all our colleagues in CSIR-NIO (RC), Kochi for their support and advice. AP and JV are grateful to Council of Scientific and Industrial Research (CSIR), New Delhi, for funding for the project (AP) and for the senior research fellowship grant (JV). Authors are grateful to Dr. Maheswari Nair, Dr. Gireesh Kumar and Mr. Muraledharan KR for the physico-chemical data and to Dr. Deepak George Pazhayamadom, Museum and Art Gallery Northern Territory (MAGNT), Darwin, Australia for IndVal analysis.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
JV is grateful to Council of Scientific and Industrial Research (CSIR), New Delhi, for the senior research fellowship grant.
Author information
Authors and Affiliations
Contributions
AP has conceptualized, designed the work, and obtained funding for research. JV and AP have collected data, analyzed samples, and prepared the manuscript. JV and NVK have performed statistical analysis of the data.
Corresponding author
Ethics declarations
Ethics approval
No need of ethics approval, since this work does not include research on identifiable human material or data.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
Rarefaction analysis of the 16S rRNA gene sequences from the bacterial communities at M1, M2, and M3 stations during PRM and MON. Fig. S2 Figure representing the taxonomic classification of the 16S rRNA gene sequences from the bacterial communities at M1, M2, and M3 stations during PRM and MON. Samples at the class level for (a) Preoteobacteria and (b) Firmicutes (DOCX 233 kb)
Rights and permissions
About this article
Cite this article
Vijayan, J., Ammini, P. & Nathan, V.K. Diversity pattern of marine culturable heterotrophic bacteria in a region with coexisting upwelling and mud banks in the southeastern Arabian Sea. Environ Sci Pollut Res 29, 3967–3982 (2022). https://doi.org/10.1007/s11356-021-15772-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-15772-8