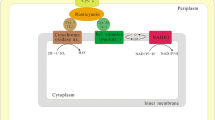
Abstract
Bacteria evolved to survive in the available environmental chemosphere via several cellular mechanisms. A rich pool of antioxidants and stress regulators plays a significant role in the survival of bacteria in unfavorable environmental conditions. Most of the microbes exhibit resistant phenomena in toxic environment niches. Naturally, bacteria possess efficient thioredoxin reductase, glutaredoxin, and peroxiredoxin redox systems to handle environmental oxidative stress. Further, an array of transcriptional regulators senses the oxidative stress conditions. Transcription regulators, such as OxyR, SoxRS, PerR, UspA, SsrB, MarA, OhrR, SarZ, etc., sense and transduce bacterial oxidative stress responses. The redox-sensitive transcription regulators continuously recycle the utilized antioxidant enzymes during oxidative stress. These regulators promote the expression of antioxidant enzymes such as superoxide dismutase, catalase, and peroxides that overcome oxidative insults. Therefore, the transcriptional regulations maintain steady-state activities of antioxidant enzymes representing the resistance against host cell/environmental oxidative insults. Further, the redox system provides reducing equivalents to synthesize biomolecules, thereby contributing to cellular repair mechanisms. The inactive transcriptional regulators in the undisturbed cells are activated by oxidative stress. The oxidized transcriptional regulators modulate the expression of antioxidant and cellular repair enzymes to survive in extreme environmental conditions. Therefore, targeting these antioxidant systems and response regulators could alter cellular redox homeostasis. This review presents the mechanisms of different redox systems that favor bacterial survival in extreme environmental oxidative stress conditions.






Similar content being viewed by others
Abbreviations
- CHP:
-
Cumene hydroperoxide
- COX:
-
Cyclooxygenase
- DNR:
-
Dissimilative nitrate respiration regulator
- FNR:
-
Fumarate-nitrate reduction regulator
- Fur:
-
Ferric uptake regulator
- γ – GCS:
-
γ glutamyl l cysteinyl glycine
- GPx:
-
Glutathione peroxidase
- GSSG:
-
Glutathione disulfide
- GS·:
-
Thiyl radical
- GR:
-
Glutathione reductase
- H2O2 :
-
Hydrogen peroxide
- HO·:
-
Hydroxyl radical
- Mrx1:
-
Mycoredoxin 1
- NsrR:
-
Nitric oxide-responsive regulator
- O2·− :
-
Superoxide anion
- ONOO− :
-
Peroxynitrite
- OHP:
-
Organic hydroperoxides
- Prx:
-
Peroxiredoxin
- RNR:
-
Ribonucleotide reductase
- -SH/-SOH:
-
Thiol-sulfenic acid
- SOD:
-
Superoxide dismutase
- Trx:
-
Thioredoxin
- TrxR:
-
Thioredoxin reductase
- UspA:
-
Universal stress protein A
References
Abbas A, Edwards C (1989) Effects of metals on a range of streptomyces species. Appl Environ Microbiol 55:2030–2035. https://doi.org/10.1128/aem.55.8.2030-2035.1989
Abbas AS, Edwards C (1990) Effects of metals on Streptomyces coelicolor growth and actinorhodin production. Appl Environ Microbiol 56:675–680. https://doi.org/10.1128/aem.56.3.675-680.1990
Abomoelak B, Hoye EA, Chi J et al (2009) mosR, a novel transcriptional regulator of hypoxia and virulence in Mycobacterium tuberculosis. J Bacteriol. https://doi.org/10.1128/JB.00778-09
Albarracín VH, Amoroso MJ, Abate CM (2005) Isolation and characterization of indigenous copper-resistant actinomycete strains. Geochemistry 65:145–156. https://doi.org/10.1016/j.chemer.2005.06.004
Albarracín VH, Pathak GP, Douki T et al (2012) Extremophilic acinetobacter strains from high-altitude lakes in Argentinean Puna: remarkable UV-B resistance and efficient DNA damage repair. Origins Life Evol Biospheres. https://doi.org/10.1007/s11084-012-9276-3
Aliyu H, De Maayer P, Cowan D (2016) The genome of the Antarctic polyextremophile Nesterenkonia sp. AN1 reveals adaptive strategies for survival under multiple stress conditions. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiw032
Alqahtani M, Ma Z, Ketkar H et al (2018) Characterization of a unique outer membrane protein required for oxidative stress resistance and virulence of Francisella tularensis. J Bacteriol. https://doi.org/10.1128/JB.00693-17
Antelmann H, Helmann JD (2011) Thiol-based redox switches and gene regulation. Antioxid Redox Signal
Argyrou A, Blanchard JS (2004) Flavoprotein disulfide reductases: advances in chemistry and function. Progr Nucleic Acid Res Mol Biol. https://doi.org/10.1016/S0079-6603(04)78003-4
Åslund F, Zheng M, Beckwith J, Storz G (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol—Disulfide status. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.96.11.6161
Atichartpongkul S, Loprasert S, Vattanaviboon P et al (2001) Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. https://doi.org/10.1099/00221287-147-7-1775. Microbiology
Badiea EA, Sayed AA, Maged M et al (2019) A novel thermostable and halophilic thioredoxin reductase from the Red Sea Atlantis II hot brine pool. PLoS ONE. https://doi.org/10.1371/journal.pone.0217565
Bano A, Hussain J, Akbar A et al (2018) Biosorption of heavy metals by obligate halophilic fungi. Chemosphere 199:218–222. https://doi.org/10.1016/j.chemosphere.2018.02.043
Barbosa TM, Pomposiello PJ (2014) The mar Regulon. Front Antimicrobial Resist
Barchiesi J, Castelli ME, Soncini FC, Véscovi EG (2008) mgtA expression is induced by Rob overexpression and mediates a Salmonella enterica resistance phenotype. J Bacteriol. https://doi.org/10.1128/JB.00195-08
Bonamore A, Gentili P, Ilari A et al (2003) Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase. J Biol Chem. https://doi.org/10.1074/jbc.M301285200
Borisov VB, Forte E, Siletsky SA et al (2015) Cytochrome bd protects bacteria against oxidative and nitrosative stress: a potential target for next-generation antimicrobial agents. Biochemistry (Moscow
Brown SM, Upadhya R, Shoemaker JD, Lodge JK (2010) Isocitrate dehydrogenase is important for nitrosative stress resistance in Cryptococcus neoformans, but oxidative stress resistance is not dependent on glucose-6-phosphate dehydrogenase. Eukaryot Cell. https://doi.org/10.1128/EC.00271-09
Brugarolas P, Movahedzadeh F, Wang Y et al (2012) The oxidation-sensing regulator (MosR) is a new Redox-dependent transcription factor in Mycobacterium tuberculosis. https://doi.org/10.2210/pdb4fx4/pdb. JBiolChem
Câmara AS, Horjales E (2018) Computer simulations reveal changes in the conformational space of the transcriptional regulator MosR upon the formation of a disulphide bond and in the collective motions that regulate its DNA-binding affinity. PLoS ONE. https://doi.org/10.1371/journal.pone.0192826
Cao Y, Jiang G, Li M et al (2020) Glutaredoxins play an important role in the redox homeostasis and symbiotic capacity of azorhizobium caulinodans ORS571. https://doi.org/10.1094/MPMI-04-20-0098-R. Molecular Plant-Microbe Interactions
Carroll RK, Liao X, Morgan LK et al (2009) Structural and functional analysis of the C-terminal DNA binding domain of the Salmonella typhimurium SPI-2 response regulator SsrB. J Biol Chem. https://doi.org/10.1074/jbc.M806261200
Castelli ME, García Véscovi E, Soncini FC (2000) The phosphatase activity is the target for Mg 2 + regulation of the sensor protein PhoQ in Salmonella. J Biol Chem. https://doi.org/10.1074/jbc.M909335199
Chaudhry GR, Mateen A, Kaskar B et al (2002) Induction of carbofuran oxidation to 4-hydroxycarbofuran by Pseudomonas sp. 50432. FEMS Microbiol Lett 214:171–176. https://doi.org/10.1016/S0378-1097(02)00851-0
Chen H, Xu G, Zhao Y et al (2008) A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS ONE. https://doi.org/10.1371/journal.pone.0001602
Chen PR, Brugarolas P, He C (2011) Redox signaling in human pathogens. Antioxid Redox Signal
Chi BK, Gronau K, Mäder U et al (2011) S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics. https://doi.org/10.1074/mcp.M111.009506
Chi BK, Busche T, Van Laer K et al (2014) Protein S-Mycothiolation functions as redox-switch and thiol protection mechanism in corynebacterium glutamicum under hypochlorite stress. Antioxid Redox Signal. https://doi.org/10.1089/ars.2013.5423
Chiang SM, Schellhorn HE (2010) Evolution of the RpoS regulon: origin of RpoS and the conservation of RpoS-dependent regulation in bacteria. J Mol Evol. https://doi.org/10.1007/s00239-010-9352-0
Chiang SM, Schellhorn HE (2012) Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Archiv Biochem Biophys
Choi C, Cho WS, Chung HK et al (2001) Prevalence of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene in isolates in weaned pigs with diarrhea and/or edema disease. Vet Microbiol. https://doi.org/10.1016/S0378-1135(01)00332-7
Cosgrove K, Coutts G, Jonsson IM et al (2007) Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. https://doi.org/10.1128/JB.01524-06
Crack JC, Green J, Thomson AJ, Brun NEL (2014) Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Acc Chem Res. https://doi.org/10.1021/ar5002507
Crawford MA, Henard CA, Tapscott T et al (2016a) DksA-dependent transcriptional regulation in Salmonella experiencing nitrosative stress. Front Microbiol. https://doi.org/10.3389/fmicb.2016.00444
Crawford MA, Tapscott T, Fitzsimmons LF et al (2016b) Redox-active sensing by bacterial DksA transcription factors is determined by cysteine and zinc content. https://doi.org/10.1128/mBio.02161-15. mBio
Dai Y, Outten FW (2012) The E. coli SufS-SufE sulfur transfer system is more resistant to oxidative stress than IscS-IscU. FEBS Lett. https://doi.org/10.1016/j.febslet.2012.10.001
Dai J, Gao K, Yao T et al (2020) Late embryogenesis abundant group3 protein (DrLEA3) is involved in antioxidation in the extremophilic bacterium Deinococcus radiodurans. Microbiol Res. https://doi.org/10.1016/j.micres.2020.126559
Dai S, Xie Z, Wang B et al (2021) Dynamic Polyphosphate Metabolism Coordinating with Manganese Ions Defends against Oxidative Stress in the Extreme Bacterium Deinococcus radiodurans. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02785-20
Daly MJ, Gaidamakova EK, Matrosova VY et al (2010a) Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE 5:10–15. https://doi.org/10.1371/journal.pone.0012570
Daly MJ, Gaidamakova EK, Matrosova VY et al (2010b) Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE. https://doi.org/10.1371/journal.pone.0012570
de Souza CS, Torres AG, Caravelli A et al (2016) Characterization of the universal stress protein F from atypical enteropathogenic Escherichia coli and its prevalence in Enterobacteriaceae. Protein Sci. https://doi.org/10.1002/pro.3038
Deponte M (2013) Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta
Ding H, Demple B (2000) Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.97.10.5146
Djorić D, Kristich CJ (2015) Oxidative stress enhances cephalosporin resistance of Enterococcus faecalis through activation of a two-component signaling system. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.03984-14
Dubbs JM, Mongkolsuk S (2016) Peroxide-sensing transcriptional regulators in bacteria. In: Stress and environmental regulation of gene expression and adaptation in bacteria
Dzink-Fox J, Oethinger M (2014) Identification of Mar mutants among clinical bacterial isolates. Front Antimicrobial Resist
El-barbary TAA, El-Badry MA (2018) Bioremediation potential of Zn (II) by different bacterial species. Saudi J Biomedical Res (SJBR) 3:144–150
El-Gebali S, Mistry J, Bateman A et al (2019) Pfam protein families database in 2019. Nucleic Acids Res
Elhosseiny NM, Amin MA, Yassin AS, Attia AS (2015) Acinetobacter baumannii universal stress protein A plays a pivotal role in stress response and is essential for pneumonia and sepsis pathogenesis. Int J Med Microbiol 305:114–123. https://doi.org/10.1016/j.ijmm.2014.11.008
El-Sayed OH, Refaat HM, Swellam MA et al (2011) Bioremediation of zinc by streptomyces aureofacienes. J Appl Sci 11:873–877. https://doi.org/10.3923/jas.2011.873.877
Ezraty B, Gennaris A, Barras F, Collet JF (2017) Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 15:385–396. https://doi.org/10.1038/nrmicro.2017.26
Fahey RC (2013) Glutathione analogs in prokaryotes. Biochim Biophys Acta
Fahey RC, Brown WC, Adams WB, Worsham MB (1978) Occurrence of glutathione in bacteria. J Bacteriol. https://doi.org/10.1128/jb.133.3.1126-1129.1978
Fang H, Chen Y, Huang L, He G (2017) Analysis of biofilm bacterial communities under different shear stresses using size-fractionated sediment. Sci Rep. https://doi.org/10.1038/s41598-017-01446-4
Fareed A, Zaffar H, Rashid A et al (2017) Biodegradation of N-methylated carbamates by free and immobilized cells of newly isolated strain Enterobacter cloacae strain TA7. Bioremediat J 21:119–127. https://doi.org/10.1080/10889868.2017.1404964
Faulkner MJ, Helmann JD (2011) Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid Redox Signal
Felix LO, Mylonakis E, Fuchs BB (2021) Thioredoxin reductase is a valid target for antimicrobial therapeutic development against Gram-Positive bacteria. Front Microbiol. https://doi.org/10.3389/fmicb.2021.663481
Friedberg EC, Aguilera A, Gellert M et al (2006) DNA repair: from molecular mechanism to human disease. In: DNA repair
Fröhlich KS, Gottesman S (2018) Small regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.rwr-0022-2018
Fuangthong M, Helmann JD (2002) The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.102483199
Fuangthong M, Helmann JD (2003) Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J Bacteriol. https://doi.org/10.1128/JB.185.21.6348-6357.2003
Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD (2001) OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol. https://doi.org/10.1128/JB.183.14.4134-4141.2001
Fuangthong M, Herbig AF, Bsat N, Helmann JD (2002) Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol. https://doi.org/10.1128/JB.184.12.3276-3286.2002
Gaballa A, Chi BK, Roberts AA et al (2014) Redox regulation in bacillus subtilis: the bacilliredoxins brxa(YPHP) and brxb(YQIW) function in de-bacillithiolation of s-bacillithiolated ohrr and mete. Antioxid Redox Signal. https://doi.org/10.1089/ars.2013.5327
Gallegos M, Michán C, Ramos JL (1993) The XylS/AraC family of regulators. Nucleic Acids Res. https://doi.org/10.1093/nar/21.4.807
García-Laviña CX, Castro-Sowinski S, Ramón A (2019) Reference genes for real-time RT-PCR expression studies in an Antarctic Pseudomonas exposed to different temperature conditions. Extremophiles 23:625–633. https://doi.org/10.1007/s00792-019-01109-4
Gaudu P, Weiss B (1996) SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.93.19.10094
Glaeser J, Klug G (2005) Photo-oxidative stress in Rhodobacter sphaeroides: protective role of carotenoids and expression of selected genes. Microbiology. https://doi.org/10.1099/mic.0.27789-0
Goodfellow M, Kämpfer P, Busse H-J et al (2012) Bergey’s manual of systematic bacteriology—volume five: the actinobacteria, Part A
Gupta J, Rathour R, Singh R, Thakur IS (2019) Production and characterization of extracellular polymeric substances (EPS) generated by a carbofuran degrading strain Cupriavidus sp. ISTL7. Bioresour Technol 282:417–424. https://doi.org/10.1016/j.biortech.2019.03.054
Handke LD, Gribenko AV, Timofeyeva Y et al (2018) MntC-dependent manganese transport is essential for Staphylococcus aureus oxidative stress resistance and virulence. https://doi.org/10.1128/msphere.00336-18. mSphere
Haugen SP, Ross W, Gourse RL (2008) Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol
Hibi T, Nii H, Nakatsu T et al (2004) Crystal structure of γ-glutamylcysteine synthetase: Insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.0403277101
Hidalgo E, Demple B (1994) An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. https://doi.org/10.1002/j.1460-2075.1994.tb06243.x
Hillmann F, Fischer RJ, Saint-Prix F et al (2008) PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.2008.06192.x
Holley CL, Zhang X, Fortney KR et al (2015) DksA and (p)ppGpp have unique and overlapping contributions to Haemophilus ducreyi pathogenesis in humans. Infect Immun. https://doi.org/10.1128/IAI.00692-15
Holmgren A (1976) Hydrogen donor system for Escherichia coli ribonucleoside diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.73.7.2275
Hsu HF, Ngo KV, Chitteni-Pattu S et al (2011) Investigating Deinococcus radiodurans RecA protein filament formation on double-stranded DNA by a real-time single-molecule approach. https://doi.org/10.1021/bi200423t. Biochemistry
Hua Y, Narumi I, Gao G et al (2003) PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. https://doi.org/10.1016/S0006-291X(03)00965-3. Biochemical and Biophysical Research Communications
Hurtado-Gallego J, Redondo-López A, Leganés F et al (2019) Peroxiredoxin (2-cys-prx) and catalase (katA) cyanobacterial-based bioluminescent bioreporters to detect oxidative stress in the aquatic environment. Chemosphere 236. https://doi.org/10.1016/j.chemosphere.2019.124395
Imlay JA (2019) Where in the world do bacteria experience oxidative. stress? Environ Microbiol
Jebeli MA, Maleki A, Amoozegar MA et al (2017) Bacillus flexus strain As-12, a new arsenic transformer bacterium isolated from contaminated water resources. Chemosphere 169:636–641. https://doi.org/10.1016/j.chemosphere.2016.11.129
Jeong SW, Choi YJ (2020) Extremophilic microorganisms for the treatment of toxic pollutants in the environment. Molecules. (Basel, Switzerland) 25
Ji Q, Zhang L, Sun F et al (2012) Staphylococcus aureus CymR is a new thiol-based oxidation-sensing regulator of stress resistance and oxidative response. J Biol Chem. https://doi.org/10.1074/jbc.M112.359737
Ji Y, Fan Y, Liu K et al (2015) Thermo activated persulfate oxidation of antibiotic sulfamethoxazole and structurally related compounds. Water Res. https://doi.org/10.1016/j.watres.2015.09.005
Jia MR, Tang N, Cao Y et al (2019) Efficient arsenate reduction by As-resistant bacterium Bacillus sp. strain PVR-YHB1-1: characterization and genome analysis. Chemosphere 218:1061–1070. https://doi.org/10.1016/j.chemosphere.2018.11.145
Kaito C, Morishita D, Matsumoto Y et al (2006) Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.2006.05480.x
Kajfasz JK, Zuber P, Ganguly T et al (2021) Increased oxidative stress tolerance of a spontaneously occurring perr gene mutation in streptococcus mutans UA159. J Bacteriol. https://doi.org/10.1128/JB.00535-20
Kalidasan V, Joseph N, Kumar S et al (2018) Iron and virulence in Stenotrophomonas Maltophilia: all we know so far. Front Cellular Infect Microbiol
Kaur S, Benov LT (2020) Methylene blue induces the soxRS regulon of Escherichia coli. https://doi.org/10.1016/j.cbi.2020.109222. Chemico-Biol Interact
Kehres DG, Maguire ME (2002) Structure, properties and regulation of magnesium transport proteins. BioMetals
Kettles RA, Tschowri N, Lyons KJ et al (2019) The Escherichia coli MarA protein regulates the ycgZ-ymgABC operon to inhibit biofilm formation. Mol Microbiol. https://doi.org/10.1111/mmi.14386
Khaleque HN, Fathollazadeh H, González C et al (2020) Unlocking survival mechanisms for metal and oxidative stress in the extremely acidophilic, halotolerant acidihalobacter genus. https://doi.org/10.3390/genes11121392. Genes
Kim SY, Lee YM (2001) Taxol-loaded block copolymer nanospheres composed of methoxy poly(ethylene glycol) and poly(ε-caprolactone) as novel anticancer drug carriers. Biomaterials. https://doi.org/10.1016/S0142-9612(00)00292-1
Kim SO, Merchant K, Nudelman R et al (2002) OxyR: A molecular code for redox-related signaling. Cell. https://doi.org/10.1016/S0092-8674(02)00723-7
Kim JS, Liu L, Vázquez-Torres A (2021) The dnak/dnaj chaperone system enables rna polymerasedksa complex formation in salmonella experiencing oxidative stress. mBio. https://doi.org/10.1128/mBio.03443-20
King M, Kubo A, Kafer L et al (2021) Calcium-regulated protein CarP responds to multiple host signals and mediates regulation of Pseudomonas aeruginosa virulence by calcium. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00061-21
Kobayashi K (2017) Sensing mechanisms in the redox-regulated, [2Fe-2S] cluster-containing, bacterial transcriptional factor SoxR. Accounts Chem Res. https://doi.org/10.1021/acs.accounts.7b00137
Kullik I, Storz G (1994) Transcriptional regulators of the oxidative stress response in prokaryotes and eukaryotes. Redox Rep. https://doi.org/10.1080/13510002.1994.11746951
Kumari N, Jagadevan S (2016) Genetic identification of arsenate reductase and arsenite oxidase in redox transformations carried out by arsenic metabolising prokaryotes—a comprehensive review. Chemosphere 163:400–412
Kvint K, Nachin L, Diez A, Nyström T (2003) The bacterial universal stress protein: function and regulation. Curr Opin Microbiol
Leelakriangsak M, Zuber P (2007) Transcription from the P3 promoter of the Bacillus subtilis spx gene is induced in response to bisulfide stress. J Bacteriol. https://doi.org/10.1128/JB.01519-06
Lehman AP, Long SR (2018) OxyRdependent transcription response of Sinorhizobium meliloti to oxidative stress. J Bacteriol. https://doi.org/10.1128/JB.00622-17
Lejona S, Castelli ME, Cabeza ML et al (2004) PhoP can activate its target genes in a PhoQ-independent manner. J Bacteriol. https://doi.org/10.1128/JB.186.8.2476-2480.2004
Lennon BW, Williams J, Ludwig ML (2000) Twists in catalysis: Alternating conformations of Escherichia coli thioredoxin reductase. Science. https://doi.org/10.1126/science.289.5482.1190
Li Y, He ZG (2012) The mycobacterial lysr-type regulator oxys responds to oxidative stress and negatively regulates expression of the catalase-peroxidase gene. PLoS ONE. https://doi.org/10.1371/journal.pone.0030186
Li XZ, Nikaido H (2009) Efflux-mediated drug resistance in bacteria: An update. Drugs
Li LH, Wu CM, Lin YT et al (2020) Roles of FadRACB system in formaldehyde detoxification, oxidative stress alleviation and antibiotic susceptibility in Stenotrophomonas maltophilia. J Antimicrob Chemother. https://doi.org/10.1093/jac/dkaa173
Li X, Liu Y, Zhong J et al (2021) Molecular mechanisms of mycoredoxin-1 in resistance to oxidative stress in corynebacterium glutamicum. J Gen Appl Microbiol. https://doi.org/10.2323/jgam.2020.03.002
Liang Q, Chiu J, Chen Y et al (2017) Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. https://doi.org/10.1158/1078-0432.CCR-16-1599
Lin YT, Huang YW, Liou RS et al (2014) MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J Antimicrob Chemother. https://doi.org/10.1093/jac/dku317
Liu X, Sun M, Cheng Y et al (2016) OxyR is a key regulator in response to oxidative stress in streptomyces avermitilis. Microbiol (United Kingdom). https://doi.org/10.1099/mic.0.000251
Lobato-Calleros C, Ramírez-Santiago C, Vernon-Carter EJ, Alvarez-Ramirez J (2014) Impact of native and chemically modified starches addition as fat replacers in the viscoelasticity of reduced-fat stirred yogurt. J Food Eng. https://doi.org/10.1016/j.jfoodeng.2014.01.019
Lu J, Holmgren A (2014) The thioredoxin antioxidant system. Free Radic Biol Med
Lu J, Vlamis-Gardikas A, Kandasamy K et al (2013) Inhibition of bacterial thioredoxin reductase: an antibiotic mechanism targeting bacteria lacking glutathione. FASEB J 27:1394–1403. https://doi.org/10.1096/fj.12-223305
Łyzén R, Maitra A, Milewska K et al (2016) The dual role of DksA protein in the regulation of Escherichia coli pArgX promoter. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw912
Ma Y, Zhang Y, Chen K et al (2021) The role of PhoP/PhoQ two component system in regulating stress adaptation in Cronobacter sakazakii. Food Microbiol. https://doi.org/10.1016/j.fm.2021.103851
Maguire ME (1992) MgtA and MgtB: prokaryotic P-type ATPases that mediate Mg2 + influx. J Bioenerg Biomembr. https://doi.org/10.1007/BF00768852
Manobala T, Shukla SK, Rao TS, Kumar MD (2021) Kinetic modelling of the uranium biosorption by Deinococcus radiodurans biofilm. Chemosphere. https://doi.org/10.1016/j.chemosphere.2020.128722
Maqbool I, Ponniresan V, Govindasamy K, Prasad NR (2019) Understanding the survival mechanisms of Deinococcus radiodurans against oxidative stress by targeting thioredoxin reductase redox system. Arch Microbiol. https://doi.org/10.1007/s00203-019-01729-6
Maqbool I, Ponniresan V, Govindasamy K, Prasad NR (2020) Understanding the survival mechanisms of Deinococcus radiodurans against oxidative stress by targeting thioredoxin reductase redox system. Arch Microbiol. https://doi.org/10.1007/s00203-019-01729-6
Martin RG, Rosner JL (1997) Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J Bacteriol. https://doi.org/10.1128/jb.179.23.7410-7419.1997
McDougald D, Gong L, Srinivasan S et al (2002) Defences against oxidative stress during starvation in bacteria. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol 81:3–13. https://doi.org/10.1023/A:1020540503200
Merino N, Aronson HS, Bojanova DP et al (2019) Living at the extremes: extremophiles and the limits of life in a planetary context. Front Microbiol 10. https://doi.org/10.3389/fmicb.2019.00780
Milse J, Petri K, Rückert C, Kalinowski J (2014) Transcriptional response of Corynebacterium glutamicum ATCC 13032 to hydrogen peroxide stress and characterization of the OxyR regulon. J Biotechnol. https://doi.org/10.1016/j.jbiotec.2014.07.452
Nachin L, Nannmark U, Nyström T (2005) Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol. https://doi.org/10.1128/JB.187.18.6265-6272.2005
Naseer N, Shapiro JA, Chander M (2014) RNA-seq analysis reveals a six-gene SoxR regulon in Streptomyces coelicolor. PLoS ONE. https://doi.org/10.1371/journal.pone.0106181
Nguyen TPO, De Mot R, Springael D (2015) Draft genome sequence of the carbofuran-mineralizing Novosphingobium sp. strain KN65.2. Genome Announc. https://doi.org/10.1128/genomeA.00764-15
Nordberg J, Arnér ESJ (2001) Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biolo Med
Nozawa R, Yokota T, Fujimoto T (1989) Susceptibility of methicillin-resistant Staphylococcus aureus to the selenium-containing compound 2-phenyl-1,2-benzoisoselenazol-3(2H)-one (PZ51). Antimicrobial Agents Chemother. https://doi.org/10.1128/AAC.33.8.1388
Ondarza RN, Rendón JL, Ondarza M (1983) Glutathione reductase in evolution. J Mol Evol. https://doi.org/10.1007/BF02101641
O’Neal CR, Gabriel WM, Turk AK et al (1994) RpoS is necessary for both the positive and negative regulation of starvation survival genes during phosphate, carbon, and nitrogen starvation in Salmonella typhimurium. J Bacteriol. https://doi.org/10.1128/jb.176.15.4610-4616.1994
Park S, Lim S, Choi J (2020) Improved tolerance of Escherichia coli to oxidative stress by expressing putative response regulator homologs from Antarctic bacteria. J Microbiol 58:131–141. https://doi.org/10.1007/s12275-020-9290-5
Parnham M, Sies H (2000) Ebselen: prospective therapy for cerebral ischaemia. Expert Opin Investig Drugs
Parshin A, Shiver AL, Lee J et al (2015) DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes sequence insertion 1. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1521365112
Patten CL, Kirchhof MG, Schertzberg MR et al (2004) Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics. https://doi.org/10.1007/s00438-004-1089-2
Perederina A, Svetlov V, Vassylyeva MN et al (2004) Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell. https://doi.org/10.1016/j.cell.2004.06.030
Pomposiello PJ, Demple B (2001) Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol 19:109–114. https://doi.org/10.1016/S0167-7799(00)01542-0
Poole LB, Nelson KJ (2008) Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol
Poor CB, Chen PR, Duguid E et al (2009) Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem. https://doi.org/10.1074/jbc.M109.015826
Qi H, Wang W, zhou, He J et al (2020) Antioxidative system of Deinococcus radiodurans. Res Microbiol
Ramanathan T, Ting YP (2016) Alkaline bioleaching of municipal solid waste incineration fly ash by autochthonous extremophiles. Chemosphere 160:54–61. https://doi.org/10.1016/j.chemosphere.2016.06.055
Ricci S, Janulczyk R, Björck L (2002) The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect Immun. https://doi.org/10.1128/IAI.70.9.4968-4976.2002
Rouhier N, Couturier J, Johnson MK, Jacquot JP (2010) Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci
Sandalova T, Zhong L, Lindqvist Y et al (2001) Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.171178698
Sato Y, Unno Y, Miyazaki C et al (2019) Multidrug-resistant Acinetobacter baumannii resists reactive oxygen species and survives in macrophages. Sci Rep. https://doi.org/10.1038/s41598-019-53846-3
Schell MA (1993) Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol
Schellhorn HE (2014) Elucidating the function of the RpoS regulon. Future Microbiol
Schellhorn HE (2020) Function, evolution, and composition of the RpoS Regulon in Escherichia coli. Front Microbiol
Schneiders T, Barbosa TM, McMurry LM, Levy SB (2004) The Escherichia coli transcriptional regulator MarA directly represses transcription of purA and hdeA. J Biol Chem. https://doi.org/10.1074/jbc.M313602200
Schröder C, Burkhardt C, Antranikian G (2020) What we learn from extremophiles. ChemTexts. https://doi.org/10.1007/s40828-020-0103-6
Seth D, Hausladen A, Stamler JS (2020) Anaerobic transcription by OxyR: a novel paradigm for nitrosative stress. Antioxid Redox Signal 32:803–816
Si M, Zhao C, Burkinshaw B et al (2017) Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1614902114
Silver S, Phung LT (1996) Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol 50:753–789
Smirnova GV, Oktyabrsky ON (2005) Glutathione in bacteria. Biokhimiya 70:1459–1473
Snavely MD, Miller CG, Maguire ME (1991) The mgtB Mg2 + transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. https://doi.org/10.1016/s0021-9258(17)35246-8
Spinelli SV, Pontel LB, García Véscovi E, Soncini FC (2008) Regulation of magnesium homeostasis in Salmonella: Mg2 + targets the mgtA transcript for degradation by RNase E. FEMS Microbiol Lett. https://doi.org/10.1111/j.1574-6968.2008.01065.x
Srijaruskul K, Charoenlap N, Namchaiw P et al (2015) Regulation by SoxR of mfsA, which encodes a major facilitator protein involved in paraquat resistance in Stenotrophomonas maltophilia. PLoS ONE. https://doi.org/10.1371/journal.pone.0123699
Storz G, Altuvia S (1994) OxyR regulon. Methods Enzymol. https://doi.org/10.1016/0076-6879(94)34088-9
Su T, Si M, Zhao Y et al (2019a) Function of alkyl hydroperoxidase AhpD in resistance to oxidative stress in Corynebacterium glutamicum. J Gen Appl Microbiol. https://doi.org/10.2323/jgam.2018.05.005
Su X, Zhou M, Hu P et al (2019b) Whole-genome sequencing of an acidophilic Rhodotorula sp. ZM1 and its phenol-degrading capability under acidic conditions. Chemosphere 232:76–86. https://doi.org/10.1016/j.chemosphere.2019.05.195
Sudharsan M, Prasad NR, Kanimozhi G et al (2022) Redox status and metabolomic profiling of thioredoxin reductase inhibitors and 4 kGy ionizing radiation-exposed Deinococcus radiodurans. Microbiol Res 261:127070. https://doi.org/10.1016/j.micres.2022.127070
Sugiman-Marangos S, Junop MS (2010) The structure of DdrB from Deinococcus: a new fold for single-stranded DNA binding proteins. Nucleic Acids Res. https://doi.org/10.1093/nar/gkq036
Sun F, Ding Y, Ji Q et al (2012) Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1205952109
Sun D, Crowell SA, Harding CM et al (2016) KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148:31–40. https://doi.org/10.1016/j.lfs.2016.02.015
Sun J, Hang Y, Han Y et al (2019) Deletion of glutaredoxin promotes oxidative tolerance and intracellular infection in Listeria monocytogenes. Virulence. https://doi.org/10.1080/21505594.2019.1685640
Tang G, Wang S, Lu D et al (2017) Two-component regulatory system ActS/ActR is required for Sinorhizobium meliloti adaptation to oxidative stress. Microbiol Res. https://doi.org/10.1016/j.micres.2017.01.005
Timková I, Sedláková-Kaduková J, Pristaš P (2018) Biosorption and bioaccumulation abilities of actinomycetes/streptomycetes isolated from metal contaminated sites. Separations 5. https://doi.org/10.3390/separations5040054
Toledano MB, Kullik I, Trinh F et al (1994) Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. https://doi.org/10.1016/S0092-8674(94)90702-1
Tsui HCT, Feng G, Winkler ME (1997) Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. https://doi.org/10.1128/jb.179.23.7476-7487.1997
Typas A, Becker G, Hengge R (2007a) The molecular basis of selective promoter activation by the σs subunit of RNA polymerase. Mol Microbiol
Typas A, Stella S, Johnson RC, Hengge R (2007b) The—35 sequence location and the Fis-sigma factor interface determine σs selectivity of the proP (P2) promoter in Escherichia coli. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.2006.05560.x
Uribe-Quero E, Rosales C (2017) Control of phagocytosis by microbial pathogens. Front Immunol 8
Vanbogelen RA, Hutton ME, Neidhardt FC (1990) Gene-Protein database of Escherichia coli K‐12: edition 3. Electrophoresis. https://doi.org/10.1002/elps.1150111205
Vázquez-Torres A (2012) Redox active thiol sensors of oxidative and nitrosative stress. Antioxid Redox Signal
Vorobjeva NV, Chernyak BV (2020) NETosis: molecular mechanisms, role in physiology and pathology. Biochemistry (Moscow)
Walthers D, Carroll RK, Navarre WW et al (2007) The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.2007.05800.x
Walthers D, Li Y, Liu Y et al (2011) Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J Biol Chem. https://doi.org/10.1074/jbc.M110.164962
Wan F, Yin J, Sun W, Gao H (2019) Oxidized OxyR up-regulates AhpCF expression to suppress plating defects of oxyR- and catalase-deficient strains. Front Microbiol 10:1–9. https://doi.org/10.3389/fmicb.2019.00439
Wang G, Maier RJ (2015) A novel DNA-binding protein plays an important role in Helicobacter pylori stress tolerance and survival in the host. J Bacteriol. https://doi.org/10.1128/JB.02489-14
Wang B, Shi Q, Ouyang Y, Chen Y (2008a) Progress in oxyR regulon- the bacterial antioxidant defense system—a review. Acta Microbiol Sin
Wang L, Li M, Dong D et al (2008b) SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J Infect Dis. https://doi.org/10.1086/586714
Wang CH, Chiang-Ni C, Kuo HT et al (2013) Peroxide responsive regulator PerR of group A streptococcus is required for the expression of phage-associated DNase Sda1 under oxidative stress. PLoS ONE. https://doi.org/10.1371/journal.pone.0081882
Wang Q, Lu X, Yang H et al (2021) Redox-sensitive transcriptional regulator SoxR directly controls antibiotic production, development and thiol-oxidative stress response in Streptomyces avermitilis. Microb Biotechnol. https://doi.org/10.1111/1751-7915.13813
Watanabe S, Kita A, Kobayashi K, Miki K (2008) Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.0709188105
Webb KM, DiRuggiero J (2013) Radiation resistance in extremophiles: fending off multiple attacks. 249–267
Xiao Y, Zhu W, He M et al (2019) High c-di-GMP promotes expression of fpr-1 and katE involved in oxidative stress resistance in Pseudomonas putida KT2440. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-019-10178-6
Xu R, Wu K, Han H et al (2018) Co-expression of YieF and PhoN in Deinococcus radiodurans R1 improves uranium bioprecipitation by reducing chromium interference. Chemosphere 211:1156–1165. https://doi.org/10.1016/j.chemosphere.2018.08.061
Yan QX, Hong Q, Han P et al (2007) Isolation and characterization of a carbofuran-degrading strain Novosphingobium sp. FND-3. FEMS Microbiol Lett 271:207–213. https://doi.org/10.1111/j.1574-6968.2007.00718.x
Yasir M, Zhang Y, Xu Z et al (2020) NAD(P)H-dependent thioredoxin-disulfide reductase TrxR is essential for tellurite and selenite reduction and resistance in Bacillus sp. Y3. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiaa126
Zapata C, Paillavil B, Chávez R et al (2017) Cytochrome c peroxidase (CcP) is a molecular determinant of the oxidative stress response in the extreme acidophilic Leptospirillum sp. CF-1. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fix001
Zheng M, Storz G (2000) Redox sensing by prokaryotic transcription factors. Biochem Pharmacol 59:1–6. https://doi.org/10.1016/S0006-2952(99)00289-0
Acknowledgements
UGC-DAE Consortium of Scientific Research has supported the work. The authors acknowledge the Department of Science and Technology (DST), New Delhi.
Funding
Funding was provided by UGC-DAE Consortium of Scientific Research (UGC-DAE-CSR-KS/CRS/19/RB-01/1044/1064).
Author information
Authors and Affiliations
Contributions
NRP conceptualized the idea and designed the work. MS carried out the computational framework and graphics and wrote the draft version of the manuscript. SR contributed to the design of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
M, S., N, R.P. & Rajendrasozhan, S. Bacterial redox response factors in the management of environmental oxidative stress. World J Microbiol Biotechnol 39, 11 (2023). https://doi.org/10.1007/s11274-022-03456-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-022-03456-5