Abstract
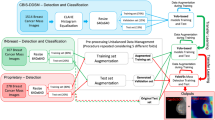
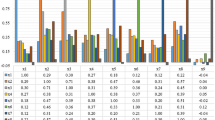
In the context of dealing with limited annotated data, this paper introduces a weakly supervised whole slide image (WSI) classification approach based on contrastive learning. The proposed method aims to detect whether cancer cells have metastasized in anterior lymph nodes of breast cancer in whole slide images. Initially, small patches are extracted from whole-slide pathology images, and an unsupervised pretraining is performed on the feature extraction model using the MoCo v2 framework. Subsequently, the feature extraction model is used to extract features from the small patches. Finally, CLAM is employed to aggregate the extracted features to obtain the overall whole slide image (WSI) classification results. Experimental results demonstrate that using MoCo v2 for unsupervised pretraining of the feature extraction model achieves an accuracy of 0.8808 in the small patch classification task. Moreover, under coarse-grained WSI-level labels, the proposed approach achieves area under the receiver operating characteristic curve (AUC) values of 0.957 ± 0.0276 and 0.9442 on different datasets, outperforming typical weakly supervised and partially supervised methods in terms of classification performance.







Similar content being viewed by others
Data Availability
Camelyon16 and Camelyon17 datasets are included in these two published article: “Bejnordi, Babak Ehteshami, et al.: Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 318.22 (2017): 2199-2210.” and “Litjens, Geert, et al.: 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience 7.6 (2018): giy065. https://doi.org/10.1093/gigascience/giy065.” 639 K-PATCH dataset is available upon request and can be obtained by contacting the corresponding author.
References
Xu WB, Zhang JD, Liu W, Lu LB et al (2021) High-precision classification method for breast cancer fusing spatial features and channel features. J Comput Appl 41(10):3025
Liu Y, Gadepalli K, Norouzi M et al (2017) Detecting cancer metastases on gigapixel pathology images. arXiv preprint arXiv:1703.02442
Eadie LH, Taylor P, Gibson AP (2012) A systematic review of computer-assisted diagnosis in diagnostic cancer imaging. Eur J Radiol 81(1):e70–e76. https://doi.org/10.1016/j.ejrad.2011.01.098
Jin X, Wen K, Lv GF et al (2020) Survey on the applications of deep learning to histopathology. J Image Graphics 25(10):1982–1993
Zhao ZX, Sun FL, Zheng S et al (2021) Research progress of artificial intelligence in the field of breast cancer histopathological diagnosis. Chin Bull Life Sci 33(05):630–637
Srinidhi CL, Ciga O, Martel AL (2021) Deep neural network models for computational histopathology: a survey. Med Image Anal 67:101813. https://doi.org/10.1016/j.media.2020.101813
Zhou X, Li C, Rahaman MM et al (2020) A comprehensive review for breast histopathology image analysis using classical and deep neural networks. IEEE Access 8:90931–90956. https://doi.org/10.1109/access.2020.2993788
Campanella G, Hanna MG, Geneslaw L et al (2019) Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 25(8):1301–1309
Qu H et al (2019) Weakly supervised deep nuclei segmentation using points annotation in histopathology images. International Conference on Medical Imaging with Deep Learning. PMLR. https://doi.org/10.1109/tmi.2020.3002244
Yang L, Zhang Y, Zhao Z et al (2018) Boxnet: deep learning based biomedical image segmentation using boxes only annotation. arXiv preprint arXiv:1806.00593
Jia Z, Huang X, Eric I et al (2017) Constrained deep weak supervision for histopathology image segmentation. IEEE Trans Med Imaging 36(11):2376–2388. https://doi.org/10.1109/tmi.2017.2724070
Lu MY, Williamson DFK, Chen TY et al (2021) Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng 5(6):555–570. https://doi.org/10.1038/s41551-020-00682-w
He K, Zhang X, Ren S et al (2016) Deep residual learning for image recognition. Proc IEEE Conf Comput Vis Pattern Recognit: 770–778. https://doi.org/10.1109/cvpr.2016.90
Russakovsky O, Deng J, Su H et al (2015) Imagenet large scale visual recognition challenge. Int J Comput Vision 115:211–252. https://doi.org/10.1007/s11263-015-0816-y
He K, Fan H, Wu Y et al (2020) Momentum contrast for unsupervised visual representation learning. Proc IEEE/CVF Conf Comput Vis Pattern Recognit: 9729–9738. https://doi.org/10.1109/cvpr42600.2020.00975
Nayak N, Chang H, Borowsky A et al (2013) Classification of tumor histopathology via sparse feature learning. 2013 IEEE 10th international symposium on biomedical imaging. IEEE, 410-413. https://doi.org/10.1109/isbi.2013.6556782
Shukla P, Verma S (2017) A compact fuzzy rule interpretation of SVM classifier for medical whole slide images. TENCON 2017–2017 IEEE Region 10 Conference. IEEE, 1588–1592. https://doi.org/10.1109/tencon.2017.8228110
Xu H, Lu C, Berendt R et al (2018) Automated analysis and classification of melanocytic tumor on skin whole slide images. Comput Med Imaging Graph 66:124–134
Klimov S, Miligy IM, Gertych A et al (2019) A whole slide image-based machine learning approach to predict ductal carcinoma in situ (DCIS) recurrence risk. Breast Cancer Res 21:1–19. https://doi.org/10.1186/s13058-019-1165-5
Courtiol P, Tramel EW, Sanselme M et al (2018) Classification and disease localization in histopathology using only global labels: a weakly-supervised approach. arXiv preprint arXiv:1802.02212
Durand T, Thome N, Cord M (2016) Weldon: weakly supervised learning of deep convolutional neural networks. Proc IEEE Conf Comput Vis Pattern Recognit: 4743–4752. https://doi.org/10.1109/cvpr.2016.513
Tellez D, van der Laak J, Ciompi F (2018) Gigapixel whole-slide image classification using unsupervised image compression and contrastive training
Kwok S (2018) Multiclass classification of breast cancer in whole-slide images. Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, June 27–29, 2018, Proceedings 15. Springer International Publishing, 931-940. https://doi.org/10.1007/978-3-319-93000-8_106
Wang S, Zhu Y, Yu L et al (2019) RMDL: recalibrated multi-instance deep learning for whole slide gastric image classification[J]. Med Image Anal 58:101549. https://doi.org/10.1016/j.media.2019.101549
Wang X, Yan Y, Tang P et al (2018) Revisiting multiple instance neural networks[J]. Pattern Recogn 74:15–24. https://doi.org/10.1016/j.patcog.2017.08.026
Li B, Li Y, Eliceiri KW (2021) Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. Proc IEEE/CVF Conf Comput Vis Pattern Recognit: 14318–14328. https://doi.org/10.1109/cvpr46437.2021.01409
Shao Z, Bian H, Chen Y et al (2021) Transmil: transformer based correlated multiple instance learning for whole slide image classification. Adv Neural Inf Process Syst 34:2136–2147
Vaswani A, Shazeer N, Parmar N et al (2017) Attention is all you need. Adv Neural Inf Process Syst 30
Oord A, Li Y, Vinyals O (2018) Representation learning with contrastive predictive coding. arXiv preprint arXiv:1807.03748
Chen T, Kornblith S, Norouzi M et al (2020) A simple framework for contrastive learning of visual representations. International conference on machine learning. PMLR: 1597–1607
Berrada L, Zisserman A, Kumar MP (2018) Smooth loss functions for deep top-k classification. arXiv preprint arXiv:1802.07595
Bejnordi BE, Veta M, Van Diest PJ et al (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22):2199–2210
Litjens G, Bandi P, EhteshamiBejnordi B et al (2018) 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience 7(6):giy065. https://doi.org/10.1093/gigascience/giy065
Bachman P, Hjelm R D, Buchwalter W (2019) Learning representations by maximizing mutual information across views. Adv Neural Inf Process Syst 32
Ma N, Zhang X, Zheng HT et al (2018) Shufflenet v2: practical guidelines for efficient cnn architecture design. Proc Eur Conf Comput Vis (ECCV): 116–131. https://doi.org/10.1007/978-3-030-01264-9_8
Xie S, Girshick R, Dollár P et al (2017) Aggregated residual transformations for deep neural networks. Proc IEEE Conf Comput Vis Pattern Recognit: 1492–1500. https://doi.org/10.1109/cvpr.2017.634
Huang G, Liu Z, Van Der Maaten L et al (2017) Densely connected convolutional networks. Proc IEEE Conf Comput Vis Pattern Recognit: 4700–4708. https://doi.org/10.1109/csci46756.2018.00084
Tellez D, Litjens G, van der Laak J et al (2019) Neural image compression for gigapixel histopathology image analysis. IEEE Trans Pattern Anal Mach Intell 43(2):567–578. https://doi.org/10.1109/tpami.2019.2936841
Dosovitskiy A, Beyer L, Kolesnikov A et al (2020) An image is worth 16x16 words: transformers for image recognition at scale[J]. arXiv preprint arXiv:2010.11929
Koohbanani NA, Qaisar T, Shaban M et al (2018) Significance of hyperparameter optimization for metastasis detection in breast histology images. Computational pathology and ophthalmic medical image analysis: first international workshop, COMPAY 2018, and 5th international workshop, OMIA 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16–20, 2018, Proceedings 5. Springer International Publishing, 139–147. https://doi.org/10.1007/978-3-030-00949-6_17
Chen X, Fan H, Girshick R et al (2020) Improved baselines with momentum contrastive learning[J]. arXiv preprint arXiv:2003.04297
Funding
This work was supported by the Harbin Science and Technology Bureau Manufacturing Innovation Talent Project (CXRC20221110393), the Heilongjiang Science and Technology Department Provincial Key R&D Program Applied Research Project (SC2022ZX06C0025), the Heilongjiang Science and Technology Department Provincial Key R&D Program Guidance Project (GZ20220088).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Yining Xie, Jun Long, Jianxin Hou, Deyun Chen and Yu Chen. The first draft of the manuscript was written by Jun Long and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Financial interests
The authors declare they have no financial interests.
Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, Y., Long, J., Hou, . et al. Weakly supervised pathological whole slide image classification based on contrastive learning. Multimed Tools Appl 83, 60809–60831 (2024). https://doi.org/10.1007/s11042-023-17988-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17988-x