Abstract
Background
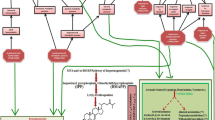
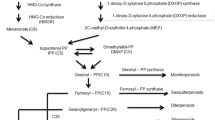
12-oxophytodienoic acid (OPDA) is a signaling molecule involved in defense and stress responses in plants. 12-oxophytodienoate reductase (OPR) is involved in the biosynthesis of jasmonic acid and trigger the conversion of OPDA into 3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0).
Methods and results
Sequence analysis revealed that Nicotiana tabacum 12-oxophytodienoate reductase 1 (OPR1) and OPR2 encoded polypeptides of 375 and 349 amino acids with molecular masses of 41.67 and 39.04 kilodaltons (kDa), respectively, while the deduced protein sequences of NtOPR1 and NtOPR2 showed high homology with other 12-oxophytodienoate reductases. BLAST (Basic local alignment search tool) analysis revealed that both NtOPRs belong to the family of Old Yellow Enzymes (OYE), and analysis of genomic DNA structure indicated that both genes include 5 exons and 4 introns. Phylogenetic analysis using MEGA X showed that NtOPR1 and NtOPR2 shared a close evolutionary relationship with Nicotiana attenuata 12-oxophytodienoate reductases. In silico analysis of subcellular localization indicated the probable locations of NtOPR1 and NtOPR2 to be the cytoplasm and the peroxisome, respectively. Tissue-specific expression assays via qRT-PCR revealed that NtOPR1 and NtOPR2 genes were highly expressed in Nicotiana tabacum roots, temperately expressed in leaves and flowers, while low expression was observed in stem tissue.
Conclusions
Presently, two 12-oxophytodienoate reductase genes (NtOPR1 and NtOPR2) were cloned and comprehensively characterized. Our findings provide comprehensive analyses that may guide future deep molecular studies of 12-oxophytodienoate reductases in Nicotiana tabacum.







Similar content being viewed by others
References
Dathe W, Rönsch H, Preiss A, Schade W, Sembdner G, Schreiber K (1981) Endogenous plant hormones of the broad bean, Vicia faba L. (-)-jasmonic acid, a plant growth inhibitor in pericarp. Planta 153:530–535
Agrawal GK, Jwa N-S, Shibato J, Han O, Iwahashi H, Rakwal R (2003) Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochem Biophys Res Commun 310:1073–1082
Ueda J, Kato J (1980) Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol 66:246–249
Gundlach H, Müller MJ, Kutchan TM, Zenk MH (1992) Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proce Nat Acad Sci 89:2389–2393
Farmer EE, Ryan CA (1990) Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proce Nat Acad Sci 87:7713–7716
Falkenstein E, Groth B, Mithöfer A, Weiler EW (1991) Methyljasmonate and α-linolenic acid are potent inducers of tendril coiling. Planta 185:316–322
Stelmach BA, Müller A, Hennig P, Laudert D, Andert L, Weiler EW (1998) Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochem 47:539–546
Jiang C, Gu J, Chopra S, Gu X, Peterson T (2004) Ordered origin of the typical two-and three-repeat Myb genes. Gene 326:13–22
Schaller F, Biesgen C, Müssig C, Altmann T, Weiler EW (2000) 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210:979–984
Engelberth J, Seidl-Adams I, Schultz JC, Tumlinson JH (2007) Insect elicitors and exposure to green leafy volatiles differentially upregulate major octadecanoids and transcripts of 12-oxo phytodienoic acid reductases in Zea mays. Mol Plant Microbe Interact 20:707–716
Costa CL, Arruda P, Benedetti CE (2000) An Arabidopsis gene induced by wounding functionally homologous to flavoprotein oxidoreductases. Plant Mole Biol 44:61–71
Nojiri H, Sugimori M, Yamane H, Nishimura Y, Yamada A, Shibuya N et al (1996) Involvement of jasmonic acid in elicitor-induced phytoalexin production in suspension-cultured rice cells. Plant Physiol 110:387–392
Vick BA, Zimmerman DC (1984) Biosynthesis of jasmonic acid by several plant species. Plant Physiol 75:458–461
Vick BA, Zimmerman DC (1986) Characterization of 12-oxo-phytodienoic acid reductase in corn: the jasmonic acid pathway. Plant Physiol 80:202–205
Dittrich H, Kutchan TM, Zenk MH (1992) The jasmonate precursor, 12-oxo-phytodienoic acid. Induces phytoalexin synthesis in Petroselinum crispum cell cultures. FEBS Lett 309:33–36
Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F (1994) The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett 345:9–13
Hamberg M (1988) Biosynthesis of 12-oxo-10, 15 (Z)-phytodienoic acid: identification of an allene oxide cyclase. Biochem Biophys Res Commun 156:543–550
Schaller F, Weiler EW (1997) Molecular Cloning and Characterization of 12-Oxophytodienoate Reductase, an Enzyme of the Octadecanoid Signaling Pathway from Arabidopsis thaliana: structural and functional relationship to yeast old yellow enzyme. J Biol Chem 272:28066–28072
Parchmann S, Gundlach H, Mueller MJ (1997) Induction of 12-oxo-phytodienoic acid in wounded plants and elicited plant cell cultures. Plant Physiol 115:1057–1064
Blechert S, Bockelmann C, Füßlein M, Tv S, Stelmach B, Niesel U et al (1999) Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207:470–479
Müssig C, Biesgen C, Lisso J, Uwer U, Weiler EW, Altmann T (2000) A novel stress-inducible 12-oxophytodienoate reductase from Arabidopsis thaliana provides a potential link between brassinosteroid-action and jasmonic-acid synthesis. J Plant Physiol 157:143–152
Sobajima H, Takeda M, Sugimori M, Kobashi N, Kiribuchi K, Cho E-M et al (2003) Cloning and characterization of a jasmonic acid-responsive gene encoding 12-oxophytodienoic acid reductase in suspension-cultured rice cells. Planta 216:692–698
Breithaupt C, Strassner J, Breitinger U, Huber R, Macheroux P, Schaller A et al (2001) X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE. Structure 9:419–429
Ishiga Y, Funato A, Tachiki T, Toyoda K, Shiraishi T, Yamada T et al (2002) Expression of the 12-oxophytodienoic acid 10, 11-reductase gene in the compatible interaction between pea and fungal pathogen. Plant Cell Physiol 43:1210–1220
Díaz M, Polanco V, Ramírez I, Peña-Cortés H (2012) Molecular cloning and expression analysis of 12-oxophytodienoate reductase cDNA by wounding in Solanum tuberosum. Electronic J Biotechnol 15:1–10
Polanco V, Ramírez I, Pena-Cortés H (2012) Molecular cloning and expression analysis of 12-oxophytodienoate reductase cDNA by wounding in Solanum tuberosum. Electronic J Biotechnol 1:2012
Rushton PJ, Bokowiec MT, Han SC, Zhang HB, Brannock JF, Chen XF, Laudeman TW, Timko MP (2008) Tobacco transcription factors: Novel insights into transcriptional regulation in the Solanaceae. Plant Physiol 147:280–295
Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5:3833
Capell T, Twyman RM, Armario-Najera V, Ma JKC, Schillberg S, Christou P (2020) Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci 25:635–643
Nogueira M, Enfissi EM, Almeida J, Fraser PD (2018) Creating plant molecular factories for industrial and nutritional isoprenoid production. Curr Opin Biotechnol 49:80–87
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Zhang J, Zhang Y, Du Y, Chen S, Tang H (2011) Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress. J Proteome Res 10:1904–1914
Ke Y, Abbas F, Zhou Y, Yu R, Fan Y (2021) Auxin-responsive R2R3-MYB transcription factors HcMYB1 and HcMYB2 activate volatile biosynthesis in Hedychium coronarium flowers. Front Plant Sci 12:710826
Ke Y, Abbas F, Zhou Y, Yu R, Yue Y, Li X et al (2019) Genome-Wide Analysis and Characterization of the Aux/IAA Family Genes Related to Floral Scent Formation in Hedychium coronarium. Inter J Mole Sci 20:3235
Liu L, Ramsay T, Zinkgraf M, Sundell D, Street NR, Filkov V et al (2015) A resource for characterizing genome-wide binding and putative target genes of transcription factors expressed during secondary growth and wood formation in Populus. Plant J 82:887–898
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biol Evol 35:1547–1549
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Stothard P (2000) The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotech 28:1102–1104
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Abbas F, Ke Y, Yu R, Fan Y (2019) Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia.’ Planta 249:71–93
Abbas F, Ke Y, Zhou Y, Ashraf U, Li X, Yu Y et al (2020) Molecular cloning, characterization and expression analysis of LoTPS2 and LoTPS4 involved in floral scent formation in oriental hybrid Lilium variety ‘Siberia.’ Phytochem 173:112294
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR et al (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY et al (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226
Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N et al (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32:585–601
Massey V (2000) The chemical and biological versatility of riboflavin. Biochem Soc Trans 28:283–296
Williams RE, Bruce NC (2002) ‘New uses for an old enzyme’–the old yellow enzyme family of flavoenzymes. Microbiol 148:1607–1614
Breithaupt C, Kurzbauer R, Lilie H, Schaller A, Strassner J, Huber R et al (2006) Crystal structure of 12-oxophytodienoate reductase 3 from tomato: self-inhibition by dimerization. Proc Nat Acad Sci 103:14337–14342
Kim S, Park J, Yeom S-I, Kim Y-M, Seo E, Kim K-T et al (2017) New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol 18:1–11
Biesgen C, Weiler E (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10, 11-reductases from Arabidopsis thaliana. Planta 208:155–165
He Y, Gan S (2001) Identical promoter elements are involved in regulation of the OPR1 gene by senescence and jasmonic acid in Arabidopsis. Plant Mole Biol 47:595–605
Beynon ER, Symons ZC, Jackson RG, Lorenz A, Rylott EL, Bruce NC (2009) The role of oxophytodienoate reductases in the detoxification of the explosive 2, 4, 6-trinitrotoluene by Arabidopsis. Plant Physiol 151:253–261
Straßner J, Fürholz A, Macheroux P, Amrhein N, Schaller A (1999) A homolog of old yellow enzyme in tomato: spectral properties and substrate specificity of the recombinant protein. J Biol Chem 274:35067–35073
Aoki K, Yano K, Suzuki A, Kawamura S, Sakurai N, Suda K et al (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genom 11:1–16
Li W, Liu B, Yu L, Feng D, Wang H, Wang J (2009) Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol Biol 9:1–19
Teif VB (2010) Predicting gene-regulation functions: lessons from temperate bacteriophages. Biophys J 98:1247–1256
de Leon S, Davidson EH (2007) Gene regulation: gene control network in development. Annu Rev Biophys Biomol Struct 36:191–212
Tani T, Sobajima H, Okada K, Chujo T, Arimura S-i, Tsutsumi N et al (2008) Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227:517–526
Mezzari MP, Walters K, Jelínkova M, Shih M-C, Just CL, Schnoor JL (2005) Gene expression and microscopic analysis of Arabidopsis exposed to chloroacetanilide herbicides and explosive compounds. A phytoremediation approach. Plant Physiol 138:858–869
Funding
This work was supported by the Science and Technology Plan Project of Yunnan Provincial Tobacco Company (2018530000241016, 2018530000241020, 2019530000241028, 2019530000241011), Shanghai Tobacco Group Co., Ltd (20193100001, 20203100001), Yunnan Provincial Department of Science and Technology (2019FH001(-051)), and Science and Technology Project of Yunnan Provincial Education Department (2021J0716).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Rights and permissions
About this article
Cite this article
Huang, F., Abbas, F., Rothenberg, D.O. et al. Molecular cloning, characterization and expression analysis of two 12-oxophytodienoate reductases (NtOPR1 and NtOPR2) from Nicotiana tabacum. Mol Biol Rep 49, 5379–5387 (2022). https://doi.org/10.1007/s11033-022-07114-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07114-9