Abstract
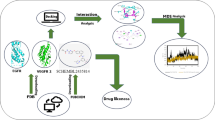
Protein tyrosine phosphatases (PTPs) are the group of enzymes that control both cellular activity and the dephosphorylation of tyrosine (Tyr)-phosphorylated proteins. Dysregulation of PTP1B has contributed to numerous diseases including Diabetes Mellitus, Alzheimer’s disease, and obesity rendering PTP1B as a legitimate target for therapeutic applications. It is highly challenging to target this enzyme because of its highly conserved and positively charged active-site pocket motivating researchers to find novel lead compounds against it. The present work makes use of an integrated approach combining ligand-based and structure-based virtual screening to find hit compounds targeting PTP1B. Initially, pharmacophore modeling was performed to find common features like two hydrogen bond acceptors, an aromatic ring and one hydrogen bond donor from the potent PTP1B inhibitors. The dataset of compounds matching with the common pharmacophoric features was filtered to remove Pan-Assay Interference substructure and to match the Lipinski criteria. Then, compounds were further prioritized using molecular docking and top fifty compounds with good binding affinity were selected for absorption, distribution, metabolism, and excretion (ADME) predictions. The top five compounds with high solubility, absorption and permeability holding score of − 10 to − 9.3 kcal/mol along with Ertiprotafib were submitted to all-atom molecular dynamic (MD) studies. The MD studies and binding free energy calculations showed that compound M4, M5 and M8 were having better binding affinity for PTP1B enzyme with ∆Gtotal score of − 24.25, − 31.47 and − 33.81 kcal/mol respectively than other compounds indicating that compound M8 could be a suitable lead compound as PTP1B inhibitor.















Similar content being viewed by others
References
Shinde RN, Kumar GS, Eqbal S, Sobhia ME (2018) Screening and identification of potential PTP1B allosteric inhibitors using in silico and in vitro approaches. PLoS ONE 13(6):e0199020. https://doi.org/10.1371/journal.pone.0199020
Tonks NK (2013) Protein tyrosine phosphatases–from housekeeping enzymes to master regulators of signal transduction. FEBS J 280(2):346–378. https://doi.org/10.1111/febs.12077
Hendriks WJ, Elson A, Harroch S, Pulido R, Stoker A, den Hertog J (2013) Protein tyrosine phosphatases in health and disease. FEBS J 280(2):708–730. https://doi.org/10.1111/febs.12000
Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A et al (2004) Protein tyrosine phosphatases in the human genome. Cell 117(6):699–711. https://doi.org/10.1016/j.cell.2004.05.018
Barford D (1996) Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem Sci 21(11):407–412. https://doi.org/10.1016/S0968-0004(96)10060-8
Ha MT, Park DH, Shrestha S, Kim M, Kim JA, Woo MH et al (2018) PTP1B inhibitory activity and molecular docking analysis of stilbene derivatives from the rhizomes of Rheum undulatum L. Fitoterapia 131:119–126. https://doi.org/10.1016/j.fitote.2018.10.020
Zhang X, Jiang H, Li W, Wang J, Cheng M (2017) Computational insight into protein tyrosine phosphatase 1b inhibition: a case study of the combined ligand-and structure-based approach. Comput Math Methods Med. https://doi.org/10.1155/2017/4245613
Johnson TO, Ermolieff J, Jirousek MR (2002) Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat Rev Drug Discov 1(9):696–709. https://doi.org/10.1038/nrd895
Ahmad F, Azevedo JL, Cortright R, Dohm GL, Goldstein BJ (1997) Alterations in skeletal muscle protein-tyrosine phosphatase activity and expression in insulin-resistant human obesity and diabetes. J Clin Investig 100(2):449–458. https://doi.org/10.1172/JCI119552
Rao PS, Muvva C, Geethanjali K, Bastipati SB, Kalashikam R (2012) Molecular docking and virtual screening for novel protein tyrosine phosphatase 1B (PTP1B) inhibitors. Bioinformation 8(17):834. https://doi.org/10.6026/97320630008834
Hsing HY, Rathnasamy S, Dianita R, Wahab HA (2020) Docking-based virtual screening in search for natural PTP1B inhibitors in treating type-2 diabetes mellitus and obesity. Biomed Res Therapy 7(1):3579–3592. https://doi.org/10.15419/bmrat.v7i1.585
Vieira MN, Lyra e Silva NM, Ferreira ST, De Felice FG, (2017) Protein tyrosine phosphatase 1B (PTP1B): a potential target for Alzheimer’s therapy? Front Aging Neurosci 9:7. https://doi.org/10.3389/fnagi.2017.00007
Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL et al (1999) Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283(5407):1544–1548. https://doi.org/10.1126/science.283.5407.1544
Cheng A, Uetani N, Simoncic PD, Chaubey VP, Lee-Loy A, McGlade CJ et al (2002) Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Dev Cell 2(4):497–503. https://doi.org/10.1016/S1534-5807(02)00149-1
Zabolotny JM, Bence-Hanulec KK, Stricker-Krongrad A, Haj F, Wang Y, Minokoshi Y et al (2002) PTP1B regulates leptin signal transduction in vivo. Dev Cell 2(4):489–495. https://doi.org/10.1016/S1534-5807(02)00148-X
Lessard L, Stuible M, Tremblay ML (2010) The two faces of PTP1B in cancer. Biochim Biophys Acta 3:613–619. https://doi.org/10.1016/j.bbapap.2009.09.018
Bollu LR, Mazumdar A, Savage MI, Brown PH (2017) Molecular pathways: targeting protein tyrosine phosphatases in cancer targeting protein tyrosine phosphatases in cancer. Clin Cancer Res 23(9):2136–2142. https://doi.org/10.1158/1078-0432.CCR-16-0934
Arias-Romero LE, Saha S, Villamar-Cruz O, Yip S-C, Ethier SP, Zhang Z-Y et al (2009) Activation of Src by protein tyrosine phosphatase 1B Is required for ErbB2 transformation of human breast epithelial cells. Can Res 69(11):4582–4588. https://doi.org/10.1158/0008-5472.CAN-08-4001
Xu Q, Wu N, Li X, Guo C, Li C, Jiang B et al (2019) Inhibition of PTP1B blocks pancreatic cancer progression by targeting the PKM2/AMPK/mTOC1 pathway. Cell Death Dis 10(12):1–15. https://doi.org/10.1038/s41419-019-2073-4
Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D (2000) Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell 6(6):1401–1412. https://doi.org/10.1016/S1097-2765(00)00137-4
Jia Z, Barford D, Flint AJ, Tonks NK (1995) Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science 268(5218):1754–1758. https://doi.org/10.1126/science.7540771
Li X, Wang L, Shi D (2016) The design strategy of selective PTP1B inhibitors over TCPTP. Bioorg Med Chem 24(16):3343–3352. https://doi.org/10.1016/j.bmc.2016.06.035
Andersen JN, Mortensen OH, GnH P, Drake PG, Iversen LF, Olsen OH et al (2001) Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol 21(21):7117–7136. https://doi.org/10.1128/MCB.21.21.7117-7136.2001
Burke TR, Lee K (2003) Phosphotyrosyl mimetics in the development of signal transduction inhibitors. Acc Chem Res 36(6):426–433. https://doi.org/10.1021/ar020127o
Ala PJ, Gonneville L, Hillman M, Becker-Pasha M, Yue EW, Douty B et al (2006) Structural insights into the design of nonpeptidic isothiazolidinone-containing inhibitors of protein-tyrosine phosphatase 1B. J Biol Chem 281(49):38013–38021. https://doi.org/10.1074/jbc.M607913200
Ala PJ, Gonneville L, Hillman MC, Becker-Pasha M, Wei M, Reid BG et al (2006) Structural basis for inhibition of protein-tyrosine phosphatase 1B by isothiazolidinone heterocyclic phosphonate mimetics. J Biol Chem 281(43):32784–32795. https://doi.org/10.1074/jbc.M606873200
Sarmiento M, Wu L, Keng Y-F, Song L, Luo Z, Huang Z et al (2000) Structure-based discovery of small molecule inhibitors targeted to protein tyrosine phosphatase 1B. J Med Chem 43(2):146–155. https://doi.org/10.1021/jm990329z
Wang Q, Huang Z, Ramachandran C, Dinaut AN, Taylor SD (1998) Naphthalenebis [α,α-difluoromethylenephosphonates] as potent inhibitors of protein tyrosine phosphatases. Bioorg Med Chem Lett 8(4):345–350. https://doi.org/10.1016/S0960-894X(98)00027-4
Taing M, Keng Y-F, Shen K, Wu L, Lawrence DS, Zhang Z-Y (1999) Potent and highly selective inhibitors of the protein tyrosine phosphatase 1B. Biochemistry 38(12):3793–3803. https://doi.org/10.1021/bi9813781
Moran EJ, Sarshar S, Cargill JF, Shahbaz MM, Lio A, Mjalli AM et al (1995) Radio frequency tag encoded combinatorial library method for the discovery of tripeptide-substituted cinnamic acid inhibitors of the protein tyrosine phosphatase PTP1B. J Am Chem Soc 117(43):10787–10788. https://doi.org/10.1021/ja00148a039
Cao L, Zhang L, Ruiz-Lozano P, Yang Q, Chien KR, Graham RM et al (1998) A novel putative protein-tyrosine phosphatase contains a BRO1-like domain and suppresses Ha-ras-mediated transformation. J Biol Chem 273(33):21077–21083. https://doi.org/10.1074/jbc.273.33.21077
Ye B, Burke TR Jr (1996) Synthesis of a difluorophosphonomethyl-containing phosphatase inhibitor designed from the X-ray structure of a PTP1B-bound ligand. Tetrahedron 52(30):9963–9970. https://doi.org/10.1016/0040-4020(96)00531-5
Burke TR, Ye B, Yan X, Wang S, Jia Z, Chen L et al (1996) Small molecule interactions with protein− tyrosine phosphatase PTP1B and their use in inhibitor design. Biochemistry 35(50):15989–15996. https://doi.org/10.1021/bi961256d
Dixit M, Saeed U, Kumar A, Siddiqi MI, Tamrakar AK, Srivastava AK, et al. (2008) Synthesis, molecular docking and PTP1B inhibitory activity of functionalized 4,5-dihydronaphthofurans and dibenzofurans. https://doi.org/10.2174/157340608783331515.
Liu S, Zeng L-F, Wu L, Yu X, Xue T, Gunawan AM et al (2008) Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors. J Am Chem Soc 130(50):17075–17084. https://doi.org/10.1021/ja8068177
Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W et al (2004) Allosteric inhibition of protein tyrosine phosphatase 1B. Nat Struct Mol Biol 11(8):730–737. https://doi.org/10.1038/nsmb803
Malamas MS, Sredy J, Gunawan I, Mihan B, Sawicki DR, Seestaller L et al (2000) New azolidinediones as inhibitors of protein tyrosine phosphatase 1B with antihyperglycemic properties. J Med Chem 43(5):995–1010. https://doi.org/10.1021/jm990476x
Wrobel J, Sredy J, Moxham C, Dietrich A, Li Z, Sawicki DR et al (1999) PTP1B inhibition and antihyperglycemic activity in the ob/ob mouse model of novel 11-arylbenzo [b] naphtho [2, 3-d] furans and 11-arylbenzo [b] naphtho [2, 3-d] thiophenes. J Med Chem 42(17):3199–3202. https://doi.org/10.1021/jm990260v
Mahapatra MK, Kumar R, Kumar M (2017) Synthesis, biological evaluation and in silico studies of 5-(3-methoxybenzylidene)thiazolidine-2,4-dione analogues as PTP1B inhibitors. Bioorg Chem 71:1–9. https://doi.org/10.1016/j.bioorg.2017.01.007
Mahapatra MK, Bera K, Singh DV, Kumar R, Kumar M (2018) In silico modelling and molecular dynamics simulation studies of thiazolidine based PTP1B inhibitors. J Biomol Struct Dyn 36(5):1195–1211. https://doi.org/10.1080/07391102.2017.1317026
Mahapatra MK, Kumar R, Kumar M (2017) N-alkylated thiazolidine-2,4-dione analogs as PTP1B inhibitors: synthesis, biological activity, and docking studies. Med Chem Res 26(6):1176–1183. https://doi.org/10.1007/s00044-017-1823-z
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44(D1):D1045–D1053. https://doi.org/10.1093/nar/gkv1072
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40(1):82–92
Sunseri J, Koes DR (2016) Pharmit: interactive exploration of chemical space. Nucleic Acids Res 44(W1):W442–W448
Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E (2021) gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput 17(10):6281–6291. https://doi.org/10.1021/acs.jctc.1c00645
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H et al (2000) The protein data bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Landrum G (2016) Rdkit: open-source cheminformatics software, 2016. URL http://www.rdkit.org/, https://github.com/rdkit/rdkit 149:150.
Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T et al (2009) KNIME-the Konstanz information miner: version 2.0 and beyond. AcM SIGKDD Explor Newslett 11(1):26–31. https://doi.org/10.1145/1656274.1656280
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminf 3(1):33. https://doi.org/10.1186/1758-2946-3-33
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791. https://doi.org/10.1002/jcc.21256
Dowarah J, Singh VP (2020) Anti-diabetic drugs recent approaches and advancements. Bioorg Med Chem 28(5):115263. https://doi.org/10.1016/j.bmc.2019.115263
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Esaki T, Ohashi R, Watanabe R, Natsume-Kitatani Y, Kawashima H, Nagao C et al (2019) Constructing an in silico three-class predictor of human intestinal absorption with Caco-2 permeability and dried-DMSO solubility. J Pharm Sci 108(11):3630–3639. https://doi.org/10.1016/j.xphs.2019.07.014
Watanabe R, Esaki T, Ohashi R, Kuroda M, Kawashima H, Komura H et al (2021) Development of an in silico prediction model for p-glycoprotein efflux potential in brain capillary endothelial cells toward the prediction of brain penetration. J Med Chem 64(5):2725–2738. https://doi.org/10.1021/acs.jmedchem.0c02011
Watanabe R, Esaki T, Kawashima H, Natsume-Kitatani Y, Nagao C, Ohashi R et al (2018) Predicting fraction unbound in human plasma from chemical structure: improved accuracy in the low value ranges. Mol Pharm 15(11):5302–5311. https://doi.org/10.1021/acs.molpharmaceut.8b00785
Esaki T, Watanabe R, Kawashima H, Ohashi R, Natsume-Kitatani Y, Nagao C et al (2019) Data curation can improve the prediction accuracy of metabolic intrinsic clearance. Mol Inf 38(1–2):1800086. https://doi.org/10.1002/minf.201800086
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Fu H, Zhang H, Chen H, Shao X, Chipot C, Cai W (2018) Zooming across the free-energy landscape: shaving barriers, and flooding valleys. J Phys Chem Lett 9(16):4738–4745. https://doi.org/10.1021/acs.jpclett.8b01994
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. https://doi.org/10.1002/jcc.20291
Ala PJ, Gonneville L, Hillman M, Becker-Pasha M, Yue EW, Douty B et al (2006) Structural insights into the design of nonpeptidic isothiazolidinone-containing inhibitors of protein-tyrosine phosphatase 1B*. J Biol Chem 281(49):38013–38021. https://doi.org/10.1074/jbc.M607913200
Wilson DP, Wan Z-K, Xu W-X, Kirincich SJ, Follows BC, Joseph-McCarthy D et al (2007) Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site. J Med Chem 50(19):4681–4698. https://doi.org/10.1021/jm0702478
Szczepankiewicz BG, Liu G, Hajduk PJ, Abad-Zapatero C, Pei Z, Xin Z et al (2003) Discovery of a potent, selective protein tyrosine phosphatase 1B inhibitor using a linked-fragment strategy. J Am Chem Soc 125(14):4087–4096. https://doi.org/10.1021/ja0296733
Xin Z, Oost TK, Abad-Zapatero C, Hajduk PJ, Pei Z, Szczepankiewicz BG et al (2003) Potent, selective inhibitors of protein tyrosine phosphatase 1B. Bioorg Med Chem Lett 13(11):1887–1890. https://doi.org/10.1016/s0960-894x(03)00302-0
Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ (2008) PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res 36(2):223–228. https://doi.org/10.1093/nar/gkn187
Scapin G, Patel SB, Becker JW, Wang Q, Desponts C, Waddleton D et al (2003) The structural basis for the selectivity of benzotriazole inhibitors of PTP1B. Biochemistry 42(39):11451–11459. https://doi.org/10.1021/bi035098j
Douty B, Wayland B, Ala PJ, Bower MJ, Pruitt J, Bostrom L et al (2008) Isothiazolidinone inhibitors of PTP1B containing imidazoles and imidazolines. Bioorg Med Chem Lett 18(1):66–71. https://doi.org/10.1016/j.bmcl.2007.11.012
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA et al (2016) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 12(1):405–413. https://doi.org/10.1021/acs.jctc.5b00935
Baidya AT, Kumar A, Uniyal A, Das B, Kumar R, Tiwari V (2021) Structure-based virtual screening and molecular dynamics simulation for the identification of sphingosine kinase-2 inhibitors as potential analgesics. J Biomol Struct Dyn 1:1–19. https://doi.org/10.1080/07391102.2021.1971559
Baidya AT, Kumar A, Kumar R, Darreh-Shori T (2022) Allosteric binding sites of Aβ peptides on the acetylcholine synthesizing enzyme ChAT as deduced by in silico molecular modeling. Int J Mol Sci 23(11):6073. https://doi.org/10.3390/ijms23116073
Evans DJ, Holian BL (1985) The nose–hoover thermostat. J Chem Phys 83(8):4069–4074. https://doi.org/10.1063/1.449071
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52(12):7182–7190. https://doi.org/10.1063/1.328693
Acknowledgements
Dr. Rajnish Kumar is grateful to the Indian Institute of Technology (BHU) Varanasi for the seed grant and Science Engineering & Research Board (SERB), India for providing start-up research grant (SRG/2021/000415). Bharti Devi is grateful to SERB for providing Junior Research Fellowship. The support and the resources provided by ‘PARAM Shivay Facility’ under the National Supercomputing Mission, Government of India at the Indian Institute of Technology (BHU), Varanasi are gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
The concept and design of the study was completed by R.K. and B.D. In silico analyses was completed by R.K., B.D., S.S.V., A.T.K, B.R.D., R.S.R., and M.K.M., R.K., B.D., S.S.V. analyzed the data. B.D. wrote the first draft of the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Devi, B., Vasishta, S.S., Das, B. et al. Integrated use of ligand and structure-based virtual screening, molecular dynamics, free energy calculation and ADME prediction for the identification of potential PTP1B inhibitors. Mol Divers 28, 649–669 (2024). https://doi.org/10.1007/s11030-023-10608-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-023-10608-8