Abstract
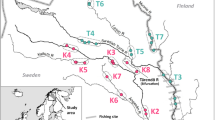
Freshwater Atlantic salmon (Salmo salar L.) populations are a living example of adaptation to the changing conditions caused by glacial cycles. The uniqueness of these populations is emphasized by almost complete resistance to the dangerous parasite Gyrodactylus salaris. In Europe, freshwater salmon populations occur primarily in north-western Russia in the republic of Karelia. These systems include Lakes Ladoga and Onega, the two largest lakes in Europe, each of which harbours a number of freshwater salmon spawning rivers. We used microsatellite markers to study the genetic structure and temporal stability of 11 freshwater salmon populations in Russian Karelia. Populations clustered according their region of origin. Although temporal variation in allele frequencies was observed in the majority of temporal comparisons, various lines of evidence demonstrated that this influence was relatively minor compared to spatial variation that explained eight times more of the variability than temporal variation. Temporal stability tended to occur in populations from rivers with a higher linear lake coefficient. The high level of genetic structuring observed in both lake systems and the apparent low level of migration between populations suggests that treating each river as a separate management unit is recommended. In addition, as the number of populations is large, the best strategy for such fine scale management would be to ensure that the level of natural reproduction in each river is sufficient to sustain the population. A prioritization strategy for population conservation based on estimating the relative roles of different evolutionary forces shaping the gene pools highlighted a number of populations where further monitoring is warranted and also identified populations which could be prioritized for conservation as living gene banks in the event that conservation resources are limited. This prioritization agreed well with the occurrence of temporal (in)stability.





Similar content being viewed by others
References
Adkinson MD (1995) Population differentiation in Pacific salmon: local adaptation, genetic drift, or the environment? Can J Fish Aquat Sci 52:2762–2777
Avise JC (2000) Phylogeography: the history and formation of species. Harward University Press, Cambridge, MA
Berg OK (1985) The formation of freshwater populations of Atlantic salmon, Salmo salar L., in Europe. J Fish Biol 27:805–815
Caballero A, Toro MA (2002) Analysis of genetic diversity for the management of conserved subdivided populations. Conserv Genet 3:289–299
Chapman MR, Shackleton NJ, Duplessy JC (2000) Sea surface temperature variability during the last glacial-interglacial cycle: assessing the magnitude and pattern of climate change in the North Atlantic. Palaeogoegr Palaeoclimatol Palaeoecol 157:1–25
Corander J, Waldmann P, Silanpää MJ (2003) Bayesian analysis of genetic differentiation between populations. Genetics 163:367–374
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater, and anadromous fishes compared with other animals. J Fish Biol 56:461–473
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Elliott SR, Coe TA, Helfield JM, Naiman RJ (1998) Spatial variation in environmental characteristics of Atlantic salmon (Salmo salar) rivers. Can J Fish Aquat Sci 55(Suppl. 1):267–280
Elphinstone MS, Hinten GN, Anderson MJ, Nock CJ (2003) An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol Ecol Notes 3:317–320
Endler JA (1986) Natural selection in the wild. Monographs in population biology 21. Princeton University Press
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fernandez J, Toro MA, Caballero A (2008) Management of subdivided populations in conservation programs: development of a novel dynamic system. Genetics 179:683–692
Fraser DJ, Hansen MM, Tessier N, Ostergaard S, Legault M, Bernatchez L (2007) Comparative estimation of effective population sizes and temporal gene flow in two contrasting population systems. Mol Ecol 16:3866–3889
Funder S, Demidov I, Yelovicheva Y (2002) Hydrography and mollusc faunas of the Baltic and the White Sea-North Sea seaway in the Eemian. Palaeogeogr Palaeoclimatol Palaeoecol 184:275–304
Garant D, Dodson JJ, Bernatchez L (2000) Ecological determinants and temporal stability of the within-river population structure in Atlantic salmon (Salmo salar L.). Mol Ecol 9:615–628
Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S, Aubin-Horth N, Lajus D, Letcher BH, Youngson AF, Webb JH, Vøllestad LA, Villanueva B, Ferguson A, Quinn TP (2007) A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol Rev 82:173–211
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Gross R, Nilsson J, Kohlman K, Lumme J, Titov S, Veselov AJe (2003) Distribution of growth hormone 1 gene haplotypes among Atlantic salmon, Salmo salar L. populations in Europe. In: Atlantic salmon: biology, conservation and restoration. Petrozavodsk, Karelian Research Centre of RAS, pp 32–37
Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361–372
Gutiérrez JP, Royo LJ, Álvarez I, Goyache F (2005) MolKin v2.0: a computer program for genetic analysis of populations using molecular coancestry information. J Heredity 96:718–721
Hansen MM, Mensberg KLD (1998) Genetic differentiation and relationship between genetic and geographical distance in Danish sea trout (Salmo trutta L.) populations. Heredity 81:493–504
Hansen MM, Ruzzante DE, Nielsen EE, Bekkevold D, Mensberg KLD (2002) Long-term effective population sizes, temporal stability of genetic composition and potential for local adaptation in anadromous brown trout (Salmo trutta) populations. Mol Ecol 11:2523–2535
Hendry AP, Letcher BH, Gries G (2003) Estimating natural selection acting on stream-dwelling Atlantic salmon: implications for the restoration of extirpated populations. Conserv Biol 17:795–805
Hewitt GM (2001) Speciation, hybrid zones and phylogeography or seeing genes in space and time. Mol Ecol 10:537–549
Hewitt GM (2004) A climate for colonization. Heredity 91:1–2
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Johnsen BO, Jensen AJ (1991) The Gyrodactylus story in Norway. Aquaculture 98:289–302
Kazakov RV (1992) Distribution of Atlantic salmon, Salmo salar L., in freshwater bodies of Europe. Aquacult Fisher Manag 23:461–475
Kazakov RV (ed) (1998) Russian salmon. Nauka, SPb (in Russian)
Kazakov RV, Titov SF (1995) Population genetic structure of Atlantic salmon. GosNIIOKhR, SPb
Kitanishi S, Yamamoto T, Higashi S (2009) Microsatellite variation reveals fine-scale genetic structure of masu salmon, Oncorhynchus masou, within the Atsuta River. Ecol Freshw Fish 18:65–71
Knutsen H, Knutsen JA, Jorde PE (2001) Genetic evidence for mixed origin of recolonized sea trout populations. Heredity 87:207–214
Koljonen ML (2001) Conservation goals and fisheries management units for Atlantic salmon in the Baltic Sea area. J Fish Biol 59(Supplement A):269–288
Kudersky LA, Ieshko E, Schulman B (2003) Distribution and range formation history of the monogenean Gyrodactylus salaris Malmberg, 1957—a parasite of juvenile Atlantic salmon Salmo salar Linnaeus, 1758. In: Veselov AJe, Ieshko EP, Nemova NN, Sterligova OP, Shustov YuA (eds) Atlantic salmon biology, conservation and restoration. Karelian Research Centre, Petrozavodsk, pp 77–83
Kuusela J, Ziętara MS, Lumme J (2007) Hybrid origin of Baltic salmon-specific parasite Gyrodactylus salaris: a model for speciation by host switch for hemiclonal organisms. Mol Ecol 16:5234–5245
Kuusela J, Holopainen R, Meinilä M, Anttila P, Koski P, Zietara MS, Veselov A, Primmer CR, Lumme J (2009) Clonal structure of salmon parasite Gyrodactylus salaris on a coevolutionary gradient on Fennoscandian salmon (Salmo salar). Ann Zool Fennici 46:21–33
Laikre L, Palm S, Ryman N (2005) Genetic population structure of fishes: implications for coastal zone management. Ambio 34:111–117
McClure MM, Holmes EE, Sanderson BL, Jordan CE (2003) A large-scale, multispecies status assignment: anadromous salmonids in the Columbia River Basin. Ecol Appl 13:964–989
Miller LM, Kapuscinski AR (1997) Historical analysis of genetic variation reveals low effective population size in a northern pike (Esox lucius) population. Genetics 147:1249–1258
Moffett IJJ, Crozier WW (1996) A study of temporal genetic variation in a natural population of Atlantic salmon in the River Bush, Northern Ireland. J Fish Biol 48:302–306
Moran P, Pendas AM, Garsia-Vasquez E, Izquierdo JI, Lobon-Cervia J (1995) Estimates of gene flow among neighbouring populations of brown trout. J Fish Biol 46:593–602
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Nielsen EE, Hansen MM, Loeschcke V (1999) Analysis of DNA from old scale samples: technical aspects, applications and perspectives for conservation. Hereditas 130:265–276
Nielsen EE, Hansen MM, Schmidt C, Meldrup D, Gronkjaer P (2001) Fisheries—population of origin of Atlantic cod. Nature 413:272
Nilsson J, Gross R, Asplund T, Dove O, Jansson H, Kelloniemi J, Kohlmann K, Löytynoja A, Nielsen EE, Paaver T, Primmer CR, Titov S, Vasemägi A, Veselov A, Öst T, Lumme J (2001) Matrilinear phylogeography of Atlantic salmon (Salmo salar L.) in Europe and postglacial colonization of the Baltic Sea area. Mol Ecol 10:89–102
Palstra FP, O’Connell MF, Ruzzante DE (2006) Temporal stability of population genetic structure of Atlantic salmon (Salmo salar) in Newfoundland. J Fish Biol 69(Supplement C):241
Palstra FP, O’Connell MF, Ruzzante DE (2007) Population structure and gene flow reversals in Atlantic salmon (Salmo salar) over contemporary and long-term temporal scales: effects of population size and life-history. Mol Ecol 16:4504–4522
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Piry S, Luikart G, Cornuet JM (1999) Bottleneck: a computer program for detecting recent reduction in the effective population size using allele frequency data. J Hered 90:502–503
Primmer CR, Veselov A, Zubchenko A, Poututkin A, Bakhmet I, Koskinen MT (2006) Isolation by distance within a river system: genetic population structuring of Atlantic salmon, Salmo salar, in tributaries of the Varzuga River in northwest Russia. Mol Ecol 15:653–666
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Richards C, Leberg PL (1996) Temporal changes in allele frequencies and a population’s history of sever bottlenecks. Conserv Biol 10:832–839
Säisä M, Koljonen ML, Gross R, Nilsson J, Tähtinen J, Koskiniemi J, Vasemägi A (2005) Population genetic structure and postglacial colonization of Atlantic salmon (Salmo salar) in the Baltic sea area based on microsatellite DNA variation. Can J Fish Aquat Sci 62:1887–1904
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schluter D (2000) The ecology of adaptive radiation. Oxford University Press, Oxford
Smirnov YuA (1971) The salmon of Lake Onega. Nauka, Leningrad (in Russian)
Swatdipong A, Vasemagi A, Koskinen M, Piironen J, Primmer CR (2009) Unanticipated population structure of European grayling in its northern distribution: implications for conservation prioritization. Front Zool 6(6) http://www.frontiersinzoology.com/content/6/I/6
Taylor EB (1991) A review of local adaptation in Salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture 98:185–207
Taylor EB, Foote CJ, Wood CC (1996) Molecular genetic evidence for parallel life-history evolution within a Pacific salmon (sockeye salmon and kokanee, Oncorhynchus nerka). Evolution 50:401–416
Tessier T, Bernatchez L (1999) Stability of population structure and genetic diversity across generations assessed by microsatellites among sympatric populations of freshwater Atlantic salmon (Salmo salar L.). Mol Ecol 8:169–179
Tessier N, Bernatchez L (2000) A genetic assessment of single versus double origin of landlocked Atlantic salmon (Salmo salar) from Lake Saint-Jean, Québec, Canada. Can J Fish Aquat Sci 57:797–804
Tonteri A, Titov S, Veselov A, Zubchenko A, Koskinen MT, Lesbarreres D, Kalyuzhin S, Bakhmet I, Lumme J, Primmer CR (2005) Phylogeography of anadromous and non-anadromous Atlantic salmon (Salmo salar) from northern Europe. Ann Zool Fennici 42:1–22
Tonteri A, Veselov AJe, Titov SI, Lumme J, Primmer CR (2007) The effect of migratory behaviour on genetic diversity and population divergence: a comparison of anadromous and freshwater Atlantic salmon Salmo salar. J Fish Biol 70:381–398
Tonteri A, Veselov AJe, Zubchenko A, Lumme J, Primmer C (2009) Microsatellites reveal clear genetic boundaries among Atlantic salmon (Salmo salar L.) populations from Barents and White seas. Can J Fish Aquat Sci 66:717–735
Toro MA, Caballero A (2005) Characterization and conservation of genetic diversity in subdivided populations. Phil Trans R Soc B 360:1367–1378
Vähä JP, Erkinaro J, Niemelä E, Primmer CR (2008) Temporally stable genetic structure and low migration in an Atlantic salmon population complex: implications for conservation and management. Evol Appl 1:137–154
Valetov VA (1999) Salmon of Lake Ladoga (biology, reproduction). Petrozavodsk (in Russian)
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vasemägi A, Gross R, Paaver T, Koljonen ML, Nilsson J (2005) Extensive immigration from compensatory hatchery releases into wild Atlantic salmon population in the Baltic sea: spatio-temporal analysis over 18 years. Heredity 95:76–83
Verspoor E, Cole LC (1989) Genetically distinct sympatric populations of resident and anadromous Atlantic salmon Salmo salar. Can J Zool 67:1453–1461
Veselov AJe (2006) Inventory and systematization of spawning rivers of the Atlantic salmon in the Murmansk oblast and Karelia. Doklady Biol Sci 407:158–162
Veselov AJe, Kalyuzhin SM (2001) Ecology, behaviour and distribution of young fish Atlantic salmon. Karelia, Petrozavodsk (in Russian)
Waples RS, Hendry AP (2008) Special issue: evolutionary perspectives on salmonid conservation and management. Evol Appl 1:183–188
Waples RS, Teel DJ (1990) Conservation genetics of Pacific salmon. I. Temporal changes in allele frequency. Conserv Biol 4:144–156
Ward RD, Woodwark M, Skibinski DOF (1994) A comparison of genetic diversity levels in marine freshwater, and anadromous fishes. J Fish Biol 44:213–232
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgements
We thank Paula Lehtonen, Jussi Kuusela, Marek Ziętara, Dmitry Stepanov, Jörg Schneider, Ville Aukee, Anni Tonteri, and Anti Vasemägi for assistance with field sampling and two anonymous reviewers for their helpful comments in improving this manuscript. This work was financed by a research program co-funded by the Finnish Academy (grant No. 124121) and Russian Foundation for Basic Researches (grant No. 08-04-91771-AF_a).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ozerov, M.Y., Veselov, A.J., Lumme, J. et al. Genetic structure of freshwater Atlantic salmon (Salmo salar L.) populations from the lakes Onega and Ladoga of northwest Russia and implications for conservation. Conserv Genet 11, 1711–1724 (2010). https://doi.org/10.1007/s10592-010-0064-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0064-1