Abstract
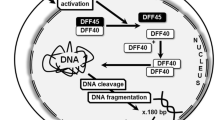
There are an increasing number of experiments to study programmed cell death/apoptosis, one of the characteristics of which is DNA fragmentation. The only current method for in situ detection of DNA fragmentation is Terminal deoxynucleotidyl transferase mediated-dUTP Nick End Labeling, TUNEL. In this study, a new method for in situ detection of apoptotic DNA fragments, namely In Situ Hybridization Chain Reaction, isHCR, was established. The principle of the assay is that the sticky end sequence of the apoptotic cell DNA fragment non-specifically initiates a hybridization chain reaction that specifically detects the apoptotic cell. The results of the combined TUNEL and isHCR method demonstrated that the majority of isHCR-positive cells were also labeled by TUNEL. In situ HCR often detect DNA fragments in the cytoplasm that the classical TUNEL method couldnot, and these cells may be in the early stages of apoptosis. It also indicates that DNA fragments are transferred to the cytoplasm during apoptosis. Because the staining process does not require terminal deoxynucleotidyl transferase as TUNEL staining does, isHCR staining cost low and can be performed on a large number of tissue specimens. It is believed that isHCR has the potential to detect DNA fragmentation of apoptotic cells in situ.




Similar content being viewed by others
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Lockshin RA, Williams CM (1965) Programmed cell death–i. cytology of degeneration in the intersegmental muscles of the Pernyi Silkmoth. J Insect Physiol 11:123–133
Maghsoudi N, Zakeri Z, Lockshin RA (2012) Programmed cell death and apoptosis–where it came from and where it is going: from Elie Metchnikoff to the control of caspases. Exp Oncol 34:146–152
Poon IK, Lucas CD, Rossi AG, Ravichandran KS (2014) Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol 14(3):166–180
Galluzzi L, Green DR (2019) Autophagy-independent functions of the autophagy machinery. Cell 177(7):1682–1699
Kerr JF, Wyllie AH, Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26:239–257
Bedoui S, Herold MJ, Strasser A (2020) Emerging connectivity of programmed cell death pathways and its physiological implications. Nat Rev Mol Cell Biol 21:678–695
Yamaguchi Y, Miura M (2015) Programmed cell death in neurodevelopment. Dev Cell 32:478–490
Elmore S (2007) Apoptosis: a review of programmed cell death. Toxicol Pathol 35:495–516
Banfalvi G (2017) Methods to detect apoptotic cell death. Apoptosis 22(2):306–323
Majtnerova P, Rousar T (2018) An overview of apoptosis assays detecting DNA fragmentation. Mol Biol Rep 45:1469–1478
Gavrieli Y, Sherman Y, Ben-Sasson SA (1992) Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol 119:493–501
Gorczyca W, Bruno S, Darzynkiewicz R, Gong J, Darzynkiewicz Z (1992) DNA strand breaks occurring during apoptosis - their early insitu detection by the terminal deoxynucleotidyl transferase and nick translation assays and prevention by serine protease inhibitors. Int J Oncol 1:639–648
Choi HM, Beck VA, Pierce NA (2014) Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8:4284–4294
Sui QQ, Zhu J, Li X, Knight GE, He C, Burnstock G, Yuan H, Xiang Z (2016) A modified protocol for the detection of three different mRNAs with a new-generation in situ hybridization chain reaction on frozen sections. J Mol Histol 47:511–529
Li F, Zhang Y, Ma SL (2016) Relationship between the expression of α-syn and neuronal apoptosis in brain cortex of acute alcoholism rats. Fa Yi Xue Za Zhi. 32(6):406–409. https://doi.org/10.3969/j.issn.1004-5619.2016.06.002
Yu Q, Guo Z, Liu X, Ouyang Q, He C, Burnstock G, Yuan H, Xiang Z (2013) Block of P2X7 receptors could partly reverse the delayed neuronal death in area CA1 of the hippocampus after transient global cerebral ischemia. Purinergic Signal 9(4):663–675
Grootjans S, Hassannia B, Delrue I et al (2016) A real-time fluorometric method for the simultaneous detection of cell death type and rate. Nat Protoc 11(8):1444–1454
Mora FAA, Musheshe N, Arroyave Ospina JC et al (2021) Metformin protects against diclofenac-induced toxicity in primary rat hepatocytes by preserving mitochondrial integrity via a pathway involving EPAC. Biomed Pharmacother 143:112072
McArthur K, Whitehead LW, Heddleston JM et al (2018) BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science. 359(6378):eaao6047
Riley JS, Quarato G, Cloix C et al (2018) Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J 37(17):e99238
Vizioli MG, Liu T, Miller KN et al (2020) Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev 34(5–6):428–445
Miller KN, Victorelli SG, Salmonowicz H et al (2021) Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell 184(22):5506–5526
Hornung V, Ablasser A, Charrel-Dennis M et al (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458(7237):514–518
Lammert CR, Frost EL, Bellinger CE et al (2020) AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment. Nature 580(7805):647–652
Funding
This work was supported by the National Natural Science Foundation of the People’s Republic of China (82171220, 81971046 to H Yuan; 818471260 to Z Xiang; 31900647 to R Ji; 81901123 to Y Lu).
Author information
Authors and Affiliations
Contributions
MY main experimental execution; analyzed the data. RJ main experimental execution; analyzed the data. ZZ main experimental execution; making animal models. WW Animal care; animal perfusion; making animal models. YL designed experiments; wrote the manuscript. ZX design the hairpins, wrote the manuscript. HY designed experiments; wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, M., Ji, R., Zhao, Z. et al. Detection of apoptotic cells based on in situ hybridization chain reaction using specific hairpins. Apoptosis 28, 222–232 (2023). https://doi.org/10.1007/s10495-022-01782-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-022-01782-5