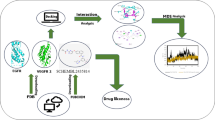
Abstract
Context
Human estrogen-related receptor γ (hERRγ) is a key protein involved in various endocrines and metabolic signaling. Numerous environmental endocrine-disrupting chemicals (EDCs) can impact related physiological activities through receptor signaling pathways. Focused on hERRγ with 4-isopropylphenol, bisphenol-F (BPF), and BP(2,2)(Un) complexes, we executed molecular docking and multiple molecular dynamics (MD) simulations along with molecular mechanics/Poisson-Boltzmann surface area (MM-PBSA) and solvation interaction energy (SIE) calculation to study the detailed dynamical structural characteristics and interactions between them. Molecular docking showed that hydrogen bonds and hydrophobic interactions were the prime interactions to keep the stability of BPF-hERRγ and hERRγ-BP(2,2)(Un) complexes. Through MD simulations, we observed that all complexes reach equilibrium during the initial 50 ns of simulation, but these three EDCs lead to local structure changes in hERRγ. Energy results further identified key residues L268, V313, L345, and F435 around the binding pockets through CH-π, π-π, and hydrogen bonds interactions play an important stabilizing role in the recognition with EDCs. And most noticeable of all, hydrophobic methoxide groups in BP(2,2)(Un) is useful for decreasing the binding ability between EDCs and hERRγ. These results may contribute to evaluate latent diseases associated with EDCs exposure at the micro level and find potential substitutes.
Method
Autodock4.2 was used to conduct the molecular docking, sietraj program was performed to calculate the energy, and VMD software was used to visualize the structure. Amber18 was conducted to perform the MD simulation and other analyses.
Graphical abstract





Similar content being viewed by others
Data availability
The datasets can be obtained from the corresponding author, through email request, reasonable request.
References
Mushtaq N, Singh DV, Bhat RA, Dervash MA, Hameed OB (2020) Freshwater contamination: sources and hazards to aquatic biota. Freshw Pollut Dynamics Remediation 27–50. https://doi.org/10.1007/978-981-13-8277-2_3
van Wezel AP, van den Hurk F, Sjerps RM, Meijers EM, Roex EW, Ter Laak TL (2018) Impact of industrial waste water treatment plants on Dutch surface waters and drinking water sources. Sci Total Environ 640:1489–1499
Yilmaz B, Terekeci H, Sandal S, Kelestimur F (2020) Endocrine disrupting chemicals: exposure, effects on human health, mechanism of action, models for testing and strategies for prevention. Rev Endocr Metab Dis 21:127–147
Sifakis S, Androutsopoulos VP, Tsatsakis AM, Spandidos DA (2017) Human exposure to endocrine disrupting chemicals: effects on the male and female reproductive systems. Environ Toxicol Phar 51:56–70
Giulivo M, de Alda ML, Capri E, Barceló D (2016) Human exposure to endocrine disrupting compounds: their role in reproductive systems, metabolic syndrome and breast cancer. A review. Environ Res 151:251–264
Thompson DC, Perera K, London R (1995) Quinone methide formation from para isomers of methylphenol (cresol), ethylphenol, and isopropylphenol: relationship to toxicity. Chem Res toxicol 8(1):55–60
Usman A, Ikhlas S, Ahmad M (2019) Occurrence, toxicity and endocrine disrupting potential of bisphenol-B and bisphenol-F: a mini-review. Toxicol Lett 312:222–227
Heard DJ, Norby PL, Holloway J, Vissing H (2000) Human ERRγ, a third member of the estrogen receptor-related receptor (ERR) subfamily of orphan nuclear receptors: tissue-specific isoforms are expressed during development and in the adult. Mol Endocrinol 14(3):382–392
Huss JM, Garbacz WG, Xie W (2015) Constitutive activities of estrogen-related receptors: transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1852(9):1912–1927
Miki K, Deguchi K, Nakanishi-Koakutsu M, Lucena-Cacace A, Kondo S, Fujiwara Y, Hatani T, Sasaki M, Naka Y, Okubo C, Narita M, Takei I, Napier SC, Sugo T, Imaichi S, Monjo T, Ando T, Tamura N, Imahashi K, Nishimoto T, Yoshida Y (2021) ERRγ enhances cardiac maturation with T-tubule formation in human iPSC-derived cardiomyocytes. Nat Commun 12(1):3596
Matsushima A, Kakuta Y, Teramoto T, Koshiba T, Liu X, Okada H, Tokunaga T, Kawabata S, Kimura M, Shimohigashi Y (2007) Structural evidence for endocrine disruptor bisphenol A binding to human nuclear receptor ERRγ. J Biochem 142(4):517–524
Thouennon E, Delfosse V, Bailly R, Blanc P, Boulahtouf A, Grimaldi M, Balaguer P (2019) Insights into the activation mechanism of human estrogen-related receptor γ by environmental endocrine disruptors. Cell Mol Life Sci 76:4769–4781
Audet-Walsh É, Yee T, McGuirk S, Vernier M, Ouellet C, St-Pierre J, Giguere V (2017) Androgen-dependent repression of ERRγ reprograms metabolism in prostate cancer role of ERRγ in prostate cancer cell metabolism. Cancer Res 77:378–389
Madhavan S, Gusev Y, Singh S, Riggins RB (2015) ERRγ target genes are poor prognostic factors in tamoxifen-treated breast cancer. J Exp Clin Canc Res 34:1–8
Fujimura T, Takahashi S, Urano T, Ijichi N, Ikeda K, Kumagai J, Mursta T, Takayama K, Horie-inoue K, Ouchi Y, Muramatsu M, Inoue S (2010) Differential expression of estrogen-related receptors β and γ (ERRβ and ERRγ) and their clinical significance in human prostate cancer. Cancer Sci 101:646–651
Legler J, Fletcher T, Govarts E, Porta M, Blumberg B, Heindel JJ, Trasande L (2015) Obesity, diabetes, and associated costs of exposure to endocrine-disrupting chemicals in the European Union. J Clin Endocrinol Metab 100:1278–1288
Amitrano A, Mahajan JS, Korley LT, Epps TH (2021) Estrogenic activity of lignin-derivable alternatives to bisphenol A assessed via molecular docking simulations. RSC Adv 11:22149–22158
Xue Q, Liu X, Liu XC, Pan WX, Fu JJ, Zhang AQ (2019) The effect of structural diversity on ligand specificity and resulting signaling differences of estrogen receptor α. Chem Res Toxicol 32:1002–1013
Li L, Wang Q, Zhang Y, Niu Y, Yao X, Liu H (2015) The molecular mechanism of bisphenol A (BPA) as an endocrine disruptor by interacting with nuclear receptors: insights from molecular dynamics (MD) simulations. PLoS ONE 10:e0120330
Sharma J, Bhardwaj VK, Das P, Purohit R (2021) Identification of naturally originated molecules as γ-aminobutyric acid receptor antagonist. J Biomol Struct Dyn 39(3):911–922
Singh R, Bhardwaj VK, Sharma J, Das P, Purohit R (2022) Identification of selective cyclin-dependent kinase 2 inhibitor from the library of pyrrolone-fused benzosuberene compounds: an in silico exploration. J Biomol Struct Dyn 40(17):7693–7701
Kumar S, Sinha K, Sharma R, Purohit R, Padwad Y (2019) Phloretin and phloridzin improve insulin sensitivity and enhance glucose uptake by subverting PPARγ/Cdk5 interaction in differentiated adipocytes. Exp Cell Res 383(1):111480
Chen L, Huang X, Li Y, Zhao B, Liang M, Wang R (2023) Structural and energetic basis of interaction between human estrogen-related receptor γ and environmental endocrine disruptors from multiple molecular dynamics simulations and free energy predictions. J Hazard Mater 443:130174
Na L, Zhou W, Yue G, Wang J, Fu W, Sun H, Li D, Duan M, Hou T (2018) Molecular dynamics simulations revealed the regulation of ligands to the interactions between androgen receptor and its coactivator. J Chem Inf Model 58:1652–1661
Sulea T, Cui Q, Purisima EO (2011) Solvated interaction energy (SIE) for scoring protein–ligand binding affinities. 2. Benchmark in the CSAR-2010 scoring exercise. J Chem Inf Model 51:2066–2081
Naïm M, Bhat S, Rankin K, Dennis S, Chowdhury S, Siddiqi I, Drabik P, Sulea T, Bayly C, Jakalian A, Purisima E (2007) Solvated interaction energy (SIE) for scoring protein−ligand binding affinities. 1. Exploring the parameter space. J Chem Inf Model 47:122–133
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Case DA, Cheatham TE III, Darden TA, Glhike H, Luo R, Merz KM, Onufriev A, Simmerling CL, Wang B, Woods RJ (2005) The amber biomolecular simualtion programs. J Comput Chem 26:1668–1688
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11:3696–3713
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Phys 25:1157–1174
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Comput Phys 79:926–935
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N.log (N) method for Ewald sums in large systems. J Comput Phys 98:10089–10092
KrUtler V, Gunsteren WFV, Hünenberger HP (2015) A fast shake algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem 22:501–508
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40(1):82–92
Hou T, Wang J, Wang Y, Li W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82
Sun H, Li Y, Tian S, Xu L, Hou T (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDB bind data set. Phys Chem Chem Phys 16:16719–16729
Swanson JM, Henchman RH, McCammon JA (2004) Revisiting free energy calculations: a theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophys J 86:67–74
Luo R, David L, Gilson MK (2002) Accelerated Poisson-Boltzmann calculations for static and dynamic systems. J Comput Chem 23:1244–1253
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Dis 10(5):449–461
Sun H, Li Y, Shen M, Tian S, Xu PP, Guan Y, Hou T (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys Chem Chem Phys 16:22035–22045
Case DA (1994) Normal mode analysis of protein dynamics. Curr Opin Struc Biol 4:285–290
Cui Q, SuleaT SJD, Munger C, Hung MN, Naïm M, Cygler ME, Purisima O (2008) Molecular dynamics-solvated interaction energy studies of protein–protein interactions: the MP1-p14 scaffolding complex. J Mol Biol 379:787–802
Hünenberger PH, Mark AE, Van Gunsteren WF (1995) Fluctuation and cross-correlation analysis of protein motions observed in nanosecond molecular dynamics simulations. J Mol Biol 252:492–503
Balsera MA, Wriggers W, Oono Y, Schulten K (1996) Principal component analysis and long time protein dynamics. J Phys Chem 100:2567–2572
Maisuradze GG, Liwo A, Scheraga HA (2009) Principal component analysis for protein folding dynamics. J Mol Biol 385:312–329
David CC, Jacobs DJ (2014) Principal component analysis: a method for determining the essential dynamics of proteins. Protein Dynamics: Methods Protoc 193–226. https://doi.org/10.1007/978-1-62703-658-0_11
Bakan A, Meireles LM, Bahar I (2011) ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27:1575–1577
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Grap Model 14:33–38
Funding
This work was supported by Fundamental Research Funds for Heilongjiang Educational Committee of Chinese hemp specialty (145209504).
Author information
Authors and Affiliations
Contributions
Lin Chen contributed to the conception of the study and wrote the manuscript; Ying Sun performed the experiment and manuscript preparation; Bing Zhao helped perform the analysis with constructive discussions; Ruige Wang performed the data analyses and wrote the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, Y., Chen, L., Zhao, B. et al. Molecular docking and molecular dynamics simulation decoding molecular mechanism of EDCs binding to hERRγ. J Mol Model 30, 127 (2024). https://doi.org/10.1007/s00894-024-05926-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-024-05926-z