Abstract
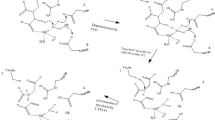
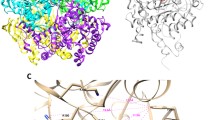
The current multidrug therapy for tuberculosis (TB) is based on the use of isoniazid (INH) in combination with other antibiotics such as rifampin, ethambutol and pyrazinamide. Literature reports have shown that Mycobacterium tuberculosis, the causative agent of TB, has become resistant to this treatment by means of point mutations in the target enzymes of these drugs, such as catalase-peroxidase (KatG). By means of equilibrium molecular dynamics in the presence of the ligand, this work evaluated ten point mutations described in the enzyme KatG that are related to resistance to INH . The results showed that the resistance mechanism is related to stereochemical modifications at the N-terminal domain of the protein, which restrict INH access to its catalytic site, not involving mechanisms of electrostatic nature. These results show insights that can be useful for the identification of new anti-TB drugs which may be able to circumvent this mechanism of resistance.





Similar content being viewed by others
References
Degen T, Bregenzer T (2016) The treatment of tuberculosis. Praxis (Bern 1994) 105(8):457–461. doi:10.1024/1661-8157/a002322
Johnsson K, King DS, Schultz PG (1995) Studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J Am Chem Soc 117:5009–5010. doi:10.1021/ja00122a038
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR Jr (1994) inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263(5144):227–230
Conde M, Fiterman J, Lima MA (2012) Tuberculosis - Sociedade Brasileira de Pneumologia e Tisiologia Grupo Gen, Rio de Janeiro
Blanchard JS (1996) Molecular mechanisms of drug resistance in mycobacterium tuberculosis. Annu Rev Biochem 65:215–239. doi:10.1146/annurev.bi.65.070196.001243
da Costa AL, Pauli I, Dorn M, Schroeder EK, Zhan CG, de Souza ON (2012) Conformational changes in 2-trans-enoyl-ACP (CoA) reductase (InhA) from M. tuberculosis induced by an inorganic complex: a molecular dynamics simulation study. J Mol Model 18(5):1779–1790. doi:10.1007/s00894-011-1200-7
Cardoso RF, Cooksey RC, Morlock GP, Barco P, Cecon L, Forestiero F, Leite CQ, Sato DN, Shikama Mde L, Mamizuka EM, Hirata RD, Hirata MH (2004) Screening and characterization of mutations in isoniazid-resistant Mycobacterium tuberculosis isolates obtained in Brazil. Antimicrob Agents Chemother 48(9):3373–3381. doi:10.1128/AAC.48.9.3373-3381.2004
Kumari R, Kumar R, Lynn A, Consort OSDD (2014) g_mmpbsa-A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962. doi:10.1021/Ci500020m
Gutierrez-de-Teran H, Aqvist J (2012) Linear interaction energy: method and applications in drug design. Methods Mol Biol 819:305–323. doi:10.1007/978-1-61779-465-0_20
Bertrand T, Eady NA, Jones JN, Jesmin NJM, Jamart-Gregoire B, Raven EL, Brown KA (2004) Crystal structure of mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279(37):38991–38999. doi:10.1074/jbc.M402382200
Vidossich P, Loewen PC, Carpena X, Fiorin G, Fita I, Rovira C (2014) Binding of the antitubercular pro-drug isoniazid in the heme access channel of catalase-peroxidase (KatG). a combined structural and metadynamics investigation. J Phys Chem B 118(11):2924–2931. doi:10.1021/jp4123425
de Beer TA, Berka K, Thornton JM, Laskowski RA (2014) PDBsum additions. Nucleic Acids Res 42:D292–296. doi:10.1093/nar/gkt940
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132. doi:10.1107/S0907444904019158
Schmidtke P, Bidon-Chanal A, Luque FJ, Barril X (2011) MDpocket: open-source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics 27(23):3276–3285. doi:10.1093/bioinformatics/btr550
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26(16):1781–1802
Mackerell AD Jr, Feig M, Brooks CL 3rd (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25(11):1400–1415. doi:10.1002/jcc.20065
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) Swiss Param: a fast force field generation tool for small organic molecules. J Comput Chem 32(11):2359–2368. doi:10.1002/jcc.21816
Neese F (2012) The ORCA program system. Wiley Interdiscip Rev: Comput Mol Sci 2(1):73–78. doi:10.1002/wcms.81
Seixas FA, Santini TD, Moura VP, Gandra EA (2008) Evaluation of the (haem)Fe--Nepsilon(2)(HisF8) bond distances from haemoglobin structures deposited in the protein data bank. Acta Crystallogr D Biol Crystallogr 64(Pt 9):971–976. doi:10.1107/S0907444908022208
Pacheco Homem D, Flores R Jr, Tosqui P, de Castro RT, Abicht Basso E, Gasparotto A Jr, Augusto Vicente Seixas F (2013) Homology modeling of dihydrofolate reductase from T. gondii bonded to antagonists: molecular docking and molecular dynamics simulations. Mol Biosyst 9(6):1308–1315. doi:10.1039/c3mb25530a
Jeeves RE, Marriott AA, Pullan ST, Hatch KA, Allnutt JC, Freire-Martin I, Hendon-Dunn CL, Watson R, Witney AA, Tyler RH, Arnold C, Marsh PD, McHugh TD, Bacon J (2015) Mycobacterium tuberculosis is resistant to isoniazid at a slow growth rate by single nucleotide polymorphisms in katG codon Ser315. PLoS One 10(9), e0138253. doi:10.1371/journal.pone.0138253
Unissa AN, Selvakumar N, Narayanan S, Suganthi C, Hanna LE (2015) Investigation of Ser315 substitutions within katG gene in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis from south India. Biomed Res Int 2015:257983. doi:10.1155/2015/257983
Zhao X, Hersleth HP, Zhu J, Andersson KK, Magliozzo RS (2013) Access channel residues Ser315 and Asp137 in Mycobacterium tuberculosis catalase-peroxidase (KatG) control peroxidatic activation of the pro-drug isoniazid. Chem Commun (Camb) 49(99):11650–11652. doi:10.1039/c3cc47022a
Suarez J, Ranguelova K, Schelvis JP, Magliozzo RS (2009) Antibiotic resistance in Mycobacterium tuberculosis: peroxidase intermediate bypass causes poor isoniazid activation by the S315G mutant of M. tuberculosis catalase-peroxidase (KatG). J Biol Chem 284(24):16146–16155. doi:10.1074/jbc.M109.005546
Zhao X, Yu H, Yu S, Wang F, Sacchettini JC, Magliozzo RS (2006) Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45(13):4131–4140. doi:10.1021/bi051967o
Purohit R, Rajendran V, Sethumadhavan R (2011) Relationship between mutation of serine residue at 315th position in M. tuberculosis catalase-peroxidase enzyme and isoniazid susceptibility: an in silico analysis. J Mol Model 17(4):869–877. doi:10.1007/s00894-010-0785-6
van Doorn HR, Kuijper EJ, van der Ende A, Welten AG, van Soolingen D, de Haas PE, Dankert J (2001) The susceptibility of Mycobacterium tuberculosis to isoniazid and the Arg--Leu mutation at codon 463 of katG are not associated. J Clin Microbiol 39(4):1591–1594. doi:10.1128/JCM.39.4.1591-1594.2001
Rodrigues L, Machado D, Couto I, Amaral L, Viveiros M (2012) Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect Genet Evol 12(4):695–700. doi:10.1016/j.meegid.2011.08.009
Acknowledgments
Fundação Araucária (convênio 147/14), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Centro Nacional de Processamento de Alto Desempenho-Sao Paulo (CENAPAD-SP) (proj512) and Laboratório Nacional de Computação Científica (LNCC).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1220 kb)
Rights and permissions
About this article
Cite this article
Pimentel, A.L., de Lima Scodro, R.B., Caleffi-Ferracioli, K.R. et al. Mutations in catalase-peroxidase KatG from isoniazid resistant Mycobacterium tuberculosis clinical isolates: insights from molecular dynamics simulations. J Mol Model 23, 121 (2017). https://doi.org/10.1007/s00894-017-3290-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-017-3290-3