Abstract
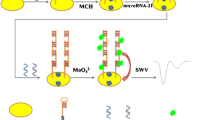
A novel and highly sensitive colorimetric DNA sensor for determination of miRNA-155 at attomolar levelsis presented that combines the peroxidase-like activity of copper nanoparticles (CuNPs) with the hybridization chain reaction (HCR) . The utilization of CuNPs offers advantages such as strong interaction with double-stranded DNA, excellent molecular recognition, and mimic catalytic activity. Herein, a capture probe DNA (P1) was immobilized on carboxylated magnetic beads (MBs), allowing for amplified immobilization due to the 3D surface. Subsequently, the presence of the target microRNA-155 led to the formation of a sandwich structure (P2/microRNA-155/P1/MBs) when P2 was introduced to the modified P1/MBs. The HCR reaction was then triggered by adding H1 and H2 to create a super sandwich (H1/H2)n. Following this, Cu2+ ions were attracted to the negatively charged phosphate groups of the (H1/H2)n and reduced by ascorbic acid, resulting in the formation of CuNPs, which were embedded into the grooves of the (H1/H2)n. The peroxidase-like activity of CuNPs catalyzed the oxidation reaction of 3,3',5,5'-Tetramethylbenzidine (TMB), resulting in a distinct blue color measured at 630 nm. Under optimal conditions, the colorimetric biosensor exhibited a linear response to microRNA–155 concentrations ranging from 80 to 500 aM, with a detection limit of 22 aM, and discriminate against other microRNAs. It was also successfully applied to the determination of microRNA–155 levels in spiked human serum.
Graphical abstract








Similar content being viewed by others
References
Chen Y-X, Huang K-J, Niu K-X (2018) Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens Bioelectron 99:612–624
Zhang Y, Yang P, Wang X-F (2014) Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol 24:153–160
Higgs G, Slack F (2013) The multiple roles of microRNA-155 in oncogenesis. J Clin Bioinforma 3:1–8
Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T (2011) Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology 79:313–320
Zadeh MM, Motamed N, Ranji N, Majidi M, Falahi F (2016) Silibinin-induced apoptosis and downregulation of microRNA-21 and microRNA-155 in MCF-7 human breast cancer cells. J Breast Cancer 19:45–52
Leng R-X, Pan H-F, Qin W-Z, Chen G-M, Ye D-Q (2011) Role of microRNA-155 in autoimmunity. Cytokine Growth Factor Rev 22:141–147
Mahesh G, Biswas R (2019) MicroRNA-155: a master regulator of inflammation. J Interferon Cytokine Res 39:321–330
Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES (2008) Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non–small cell lung cancer by quantitative real-time RT-PCR. Clin Chem 54:1696–1704
Moustakim H, Mohammadi H, Amine A (2022) Electrochemical DNA biosensor based on immobilization of a non-modified ssDNA using phosphoramidate-bonding strategy and pencil graphite electrode modified with AuNPs/CB and self-assembled cysteamine monolayer. Sensors 22:9420
Chahri I, Karrat A, Mohammadi H, Amine A (2023) Development of a new route for the immobilization of unmodified single-stranded DNA on chitosan beads and detection of released guanine after hydrolysis. Molecules 28:2088
El Aamri M, Yammouri G, Mohammadi H, Amine A, Korri-Youssoufi H (2020) Electrochemical biosensors for detection of microRNA as a cancer biomarker: Pros and cons. Biosensors 10:186
El Aamri M, Mohammadi H, Amine A (2023) Development of a novel electrochemical sensor based on functionalized carbon black for the detection of guanine released from DNA hydrolysis. Electroanalysis 35:e202100613
Wang Y, Yang Q, Gao Z, Dong H (2022) Recent advance of RNA aptamers and DNAzymes for MicroRNA detection. Biosens Bioelectron 212:114423
Aamri ME, Mohammadi H, Amine A (2022) Novel label-free colorimetric and electrochemical detection for MiRNA-21 based on the complexation of molybdate with phosphate. Microchem J 182:107851. https://doi.org/10.1016/j.microc.2022.107851
Mohammadi H, Amine A (2018) Spectrophotometric and electrochemical determination of MicroRNA-155 using sandwich hybridization magnetic beads. Anal Lett 51:411–423
Lamprou E, Sotiriou M, Kalligosfyri PM, Kalogianni DP, Christopoulos TK (2023) A universal lateral flow assay for microRNA visual detection in urine samples. Talanta 262:124682
Mohammadi H, Yammouri G, Amine A (2019) Current advances in electrochemical genosensors for detecting microRNA cancer markers. Curr Opin Electrochem 16:96–105. https://doi.org/10.1016/j.coelec.2019.04.030
Zouari M, Campuzano S, Pingarrón JM, Raouafi N (2020) Femtomolar direct voltammetric determination of circulating miRNAs in sera of cancer patients using an enzymeless biosensor. Anal Chim Acta 1104:188–198
Baachaoui S, Mastouri M, Meftah M, Yaacoubi-Loueslati B, Raouafi N (2023) A Magnetoelectrochemical bioassay for highly sensitive sensing of point mutations in Interleukin-6 gene using TMB as a hybridization intercalation indicator. Biosensors 13:240. https://doi.org/10.3390/bios13020240
Granados-Riveron JT, Aquino-Jarquin G (2021) CRISPR/Cas13-based approaches for ultrasensitive and specific detection of microRNAs. Cells 10:1655
Liu J, Xie G, Lv S, Xiong Q, Xu H (2023) Recent applications of rolling circle amplification in biosensors and DNA nanotechnology. TrAC Trends Anal Chem 160:116953
Gao Y, Huang K, Wang F, Hou Y, Wang B, Xu J, Shuai H, Li G (2023) The self-powered electrochemical biosensing platform with multi-amplification strategy for ultrasensitive detection of microRNA-155. Anal Chim Acta 1239:340702
Weng S, Lin D, Lai S, Tao H, Chen T, Peng M, Qiu S, Feng S (2022) Highly sensitive and reliable detection of microRNA for clinically disease surveillance using SERS biosensor integrated with catalytic hairpin assembly amplification technology. Biosens Bioelectron 208:114236
Luo Z, Li Y, Zhang P, He L, Feng Y, Feng Y, Qian C, Tian Y, Duan Y (2022) Catalytic hairpin assembly as cascade nucleic acid circuits for fluorescent biosensor: design, evolution and application. TrAC Trends Anal Chem 151:116582
Mohan B, Kumar S, Kumar S, Modi K, Tyagi D, Papukashvili D, Rcheulishvili N, Pombeiro AJ (2023) Nanomaterials for miRNA detection: the hybridization chain reaction strategy. Sens Diagn 2:78–89
Gao G, Hu J, Li Z, Xu Q, Wang C-S, Jia H-M, Zhou H, Lin P, Zhao W-W (2022) Hybridization chain reaction for regulating surface capacitance of organic photoelectrochemical transistor toward sensitive miRNA detection. Biosens Bioelectron 209:114224
Ge Z, Lin M, Wang P, Pei H, Yan J, Shi J, Huang Q, He D, Fan C, Zuo X (2014) Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal Chem 86:2124–2130
Gao X, Xu L-P, Wu T, Wen Y, Ma X, Zhang X (2016) An enzyme-amplified lateral flow strip biosensor for visual detection of microRNA-224. Talanta 146:648–654
Mandli J, Mohammadi H, Amine A (2017) Electrochemical DNA sandwich biosensor based on enzyme amplified microRNA-21 detection and gold nanoparticles. Bioelectrochemistry 116:17–23
Gao L, Zhuang J, Nie L, Zhang J, Zhang Y, Gu N, Wang T, Feng J, Yang D, Perrett S (2007) Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat Nanotechnol 2:577–583
Aamri ME, Mohammadi H, Amine A (2023) Paper-based colorimetric detection of miRNA-21 using pre-activated nylon membrane and peroxidase-mimetic activity of cysteamine-capped gold nanoparticles. Biosensors 13:74. https://doi.org/10.3390/bios13010074
Wang H, Wan K, Shi X (2019) Recent advances in nanozyme research. Adv Mater 31:1805368
Robert A, Meunier B (2022) How to define a nanozyme. ACS Nano 16:6956–6959
Wu J, Lv W, Yang Q, Li H, Li F (2021) Label-free homogeneous electrochemical detection of MicroRNA based on target-induced anti-shielding against the catalytic activity of two-dimension nanozyme. Biosens Bioelectron 171:112707
Li Y, Zhang C, He Y, Gao J, Li W, Cheng L, Sun F, Xia P, Wang Q (2022) A generic and non-enzymatic electrochemical biosensor integrated molecular beacon-like catalyzed hairpin assembly circuit with MOF@ Au@ G-triplex/hemin nanozyme for ultrasensitive detection of miR-721. Biosens Bioelectron 203:114051
Tian L, Qi J, Oderinde O, Yao C, Song W, Wang Y (2018) Planar intercalated copper (II) complex molecule as small molecule enzyme mimic combined with Fe3O4 nanozyme for bienzyme synergistic catalysis applied to the microRNA biosensor. Biosens Bioelectron 110:110–117
Liu Y, Zheng Y, Ding D, Guo R (2017) Switching peroxidase-mimic activity of protein stabilized platinum nanozymes by sulfide ions: substrate dependence, mechanism, and detection. Langmuir 33:13811–13820
Wu Y, Chen W, Wang C, Xing D (2023) Overview of nanozymes with phosphatase-like activity. Biosens Bioelectron 237:115470
Huang Y, Liu Z, Liu C, Ju E, Zhang Y, Ren J, Qu X (2016) Self-assembly of multi-nanozymes to mimic an intracellular antioxidant defense system. Angew Chem 128:6758–6762
Golchin J, Golchin K, Alidadian N, Ghaderi S, Eslamkhah S, Eslamkhah M, Akbarzadeh A (2017) Nanozyme applications in biology and medicine: an overview. Artif Cells Nanomed Biotechnol 45:1069–1076
Qing Z, Bai A, Xing S, Zou Z, He X, Wang K, Yang R (2019) Progress in biosensor based on DNA-templated copper nanoparticles. Biosens Bioelectron 137:96–109
Liu R, Wang C, Hu J, Su Y, Lv Y (2018) DNA-templated copper nanoparticles: versatile platform for label-free bioassays. TrAC Trends Anal Chem 105:436–452
Chen C-A, Chen S-C, Shiddiky MJ, Chen C-F, Wu KC-W (2020) DNA-templated copper nanoprobes: overview, feature, application, and current development in detection technologies. Chem Rec 20:174–186
Cao Q, Li J, Wang E (2019) Recent advances in the synthesis and application of copper nanomaterials based on various DNA scaffolds. Biosens Bioelectron 132:333–342
Alipour M, Jalili S, Shirzad H, Ansari Dezfouli E, Fouani MH, Sadeghan AA, Bardania H, Hosseinkhani S (2020) Development of dual-emission cluster of Ag atoms for genetically modified organisms detection. Microchim Acta 187:628. https://doi.org/10.1007/s00604-020-04591-2
Miao P, Zhang T, Xu J, Tang Y (2018) Electrochemical detection of miRNA combining T7 exonuclease-assisted cascade signal amplification and DNA-templated copper nanoparticles. Anal Chem 90:11154–11160
Xu F, Shi H, He X, Wang K, He D, Guo Q, Qing Z, Yan L, Ye X, Li D (2014) Concatemeric dsDNA-templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in microRNA detection. Anal Chem 86:6976–6982
Vargas E, Povedano E, Montiel VR-V, Torrente-Rodríguez RM, Zouari M, Montoya JJ, Raouafi N, Campuzano S, Pingarrón JM (2018) Single-step incubation determination of miRNAs in cancer cells using an amperometric biosensor based on competitive hybridization onto magnetic beads. Sensors 18:863
Wang Y, Meng W, Chen X, Zhang Y (2020) DNA-templated copper nanoparticles as signalling probe for electrochemical determination of microRNA-222. Microchim Acta 187:1–9
Liao H, Hu L, Zhang Y, Yu X, Liu Y, Li R (2018) A highly selective colorimetric sulfide assay based on the inhibition of the peroxidase-like activity of copper nanoclusters. Microchim Acta 185:1–6
Kaur P, Thakur R, Chaudhury A (2016) Biogenesis of copper nanoparticles using peel extract of Punica granatum and their antimicrobial activity against opportunistic pathogens. Green Chem Lett Rev 9:33–38
Tang Y, Liu M, Zhao Z, Li Q, Liang X, Tian J, Zhao S (2019) Fluorometric determination of microRNA-122 by using ExoIII-aided recycling amplification and polythymine induced formation of copper nanoparticles. Microchim Acta 186:133. https://doi.org/10.1007/s00604-019-3237-8
Chi B-Z, Liang R-P, Qiu W-B, Yuan Y-H, Qiu J-D (2017) Direct fluorescence detection of microRNA based on enzymatically engineered primer extension poly-thymine (EPEPT) reaction using copper nanoparticles as nano-dye. Biosens Bioelectron 87:216–221. https://doi.org/10.1016/j.bios.2016.08.042
Shuofeng L, Fangfang W, Wang C, Wang Z, Wu Q (2023) Sensitive detection of microRNA based on high-fidelity CRISPR/Cas13a trans cleavage activity coupled with template-free DNA extension-induced strongly emitting copper nanoparticles. Sens Actuators B Chem 374:132848. https://doi.org/10.1016/j.snb.2022.132848
Xu F, Qiao Z, Luo L, He X, Lei Y, Tang J, Shi H, Wang K (2022) A label-free cyclic amplification strategy for microRNA detection by coupling graphene oxide-controlled adsorption with superlong poly(thymine)-hosted fluorescent copper nanoparticles. Talanta 243:123323. https://doi.org/10.1016/j.talanta.2022.123323
Xu F, Luo L, Shi H, He X, Lei Y, Tang J, He D, Qiao Z, Wang K (2018) Label-free and sensitive microRNA detection based on a target recycling amplification-integrated superlong poly(thymine)-hosted copper nanoparticle strategy. Anal Chim Acta 1010:54–61. https://doi.org/10.1016/j.aca.2018.01.010
Acknowledgements
The authors acknowledge the Moroccan Ministry of Higher Education, Scientific Research and Innovation and the OCP Foundation « APRD research program 2021».
Funding
This research was funded by the Moroccan Ministry of Higher Education, Scientific Research and Innovation and the OCP Foundation « APRD research program 2021».
Author information
Authors and Affiliations
Contributions
Maliana El Aamri: Methodology, Fabrication, Data Curation, Investigation, Writing-Original draft preparation. Hasna Mohammadi: Writing-Review & Editing, Review & Editing, Supervision, funding acquisition. Aziz Amine: Writing-Review & Editing, Review & Editing, Supervision, funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• An original non-enzymatic bio-assay was developed for an attomolar-level colorimetric microRNA-155 detection.
• The ingenious combination of HCR with the peroxidase-like activity of copper nanoparticles enhances sensitivity while concurrently lowering the detection limit.
• An ultralow detection limit of 22 aM was achieved for the direct detection of microRNA-155 biomarker in human serum samples.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
EL Aamri, M., Mohammadi, H. & Amine, A. A highly sensitive colorimetric DNA sensor for MicroRNA-155 detection: leveraging the peroxidase-like activity of copper nanoparticles in a double amplification strategy. Microchim Acta 191, 32 (2024). https://doi.org/10.1007/s00604-023-06087-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-06087-1