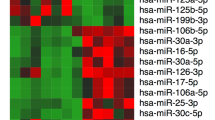
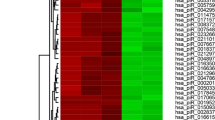
Abstract
Many studies have shown that circRNAs and miRNAs play important roles in many different life processes. However, the function of circRNAs in spermatogenesis remains unknown. Here, we aimed to explore the mechanisms whereby circRNA-miRNAs-mRNAs regulate abnormal m6A methylation in GC-1spg spermatogonia. We first reduced m6A methylation in GC-1spg whole cells after knocking down the m6A methyltransferase enzyme, METTL3. Then, we performed circRNA- and miRNA-seq on GC-1spg cells with low m6A methylation and identified 48 and 50 differentially expressed circRNAs and miRNAs, respectively. We also predicted the targets of the differentially expressed miRNAs by using Miranda software and further constructed the differentially expressed circRNA-differentially expressed miRNA-mRNA ceRNA network. GO analysis was performed on the differentially expressed circRNAs and miRNA-targeted mRNAs, and an interaction network between the proteins of interest was constructed using Cytoscape. The final GO analysis showed that the target mRNAs were involved in sperm formation. Therefore, a PPI network was subsequently constructed and 2 hub genes (H2afx and Dnmt3a) were identified. In this study, we constructed a ceRNA network and explored the regulatory roles of circRNAs and miRNAs in the pathogenesis of abnormal spermatogenesis caused by low levels of methylated m6A. Also, we identified two pivotal genes that may be key factors in infertility caused by abnormal m6A methylation. This may provide some ideas for the treatment of infertility resulting from abnormal spermatogenesis.









Similar content being viewed by others
Data availability
Available underlying data will be provided upon reasonable request.
References
Ambros V (2003) Microrna pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113(6):673–676
Cao S, Wang G, Wang J, Li C, Zhang L (2019) Hsa_circ_101280 promotes hepatocellular carcinoma by regulating mir-375/jak2. Immunol Cell Biol 97(2):218–228
Carrell DT (2012) Epigenetics of the male gamete. Fertil Steril 97(2):267–274
Chen C, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science (New York, N.Y.), 303(5654), 83–86
Chen L, Yang L (2015) Regulation of Circrna Biogenesis Rna Biol 12(4):381–388
Chen X, Li X, Guo J, Zhang P, Zeng W (2017) The roles of microRNAs in regulation of mammalian spermatogenesis. J Anim Sci Biotechno 8:35
Chen Z, Zhang Y (2020) Role of mammalian DNA methyltransferases in development. Annu Rev Biochem 89:135–158
Chioccarelli T, Manfrevola F, Ferraro B, Sellitto C, Cobellis G, Migliaccio M et al (2019) Expression patterns of circular RNAs in high quality and poor quality human spermatozoa. Front Endocrinol 10:435
Chioccarelli T, Pierantoni R, Manfrevola F, Porreca V, Fasano S, Chianese R et al (2020) Histone post-translational modifications and circRNAs in mouse and human spermatozoa: potential epigenetic marks to assess human sperm quality. J Clin Med 9(3):640
Dada R, Kumar M, Jesudasan R, Fernández J, Gosálvez L, J. Agarwal A. (2012) Epigenetics and its role in male infertility. J Assist Reprod Gen 29(3):213–223
de Rooij DG (2001) Proliferation and differentiation of spermatogonial stem cells. Reproduction (cambridge, England) 121(3):347–354
Desimio MG, Cesari E, Sorrenti M, De Felici M, Farini D (2021) Stimulated by retinoic acid gene 8 (STRA8) interacts with the germ cell specific bHLH factor SOHLH1 and represses c-KIT expression in vitro. J Cell Mol Med 25(1):383–396
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S (2012) Topology of the human and mouse m6a RNA methylomes revealed by m6a-seq., (Vol. 485,201–206
Dura M, Teissandier A, Armand M, Barau J, Lapoujade C, Fouchet P et al (2022) DNMT3A-dependent DNA methylation is required for spermatogonial stem cells to commit to spermatogenesis. Nat Genet 54(4):469–480
Fernandez-Capetillo O, Mahadevaiah S, Celeste K, A. Romanienko P. J. Camerini-Otero R. D. Bonner W. M. et al (2003) H2ax is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4(4):497–508
Finocchi F, Pelloni M, Balercia G, Pallotti F, Radicioni A, F. Lenzi A. et al (2020) Seminal plasma miRNAs in Klinefelter syndrome and in obstructive and non-obstructive azoospermia. Mol Biol Rep 47(6):4373–4382
Ge P, Zhang J, Zhou L, Lv M, Li Y, Wang J et al (2019) CircRNA expression profile and functional analysis in testicular tissue of patients with non-obstructive azoospermia. Reproductive Biology and Endocrinology : RB&E 17(1):100
Griswold MD (2016) Spermatogenesis: the commitment to meiosis. Physiol Rev 96(1):1–17
Gui Y, Yuan S (2021) Epigenetic regulations in mammalian spermatogenesis: RNA-m(6)a modification and beyond. Cellular and Molecular Life Sciences : CMLS 78(11):4893–4905
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Hayashi K, de Sousa C, Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K et al (2008) MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 3(3):e1738
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y et al (2017) Ythdc2 is an n(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27(9):1115–1127
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, N.Y.), 19(2), 141–157
Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E et al (2004) Essential role for de novo DNA methyltransferase dnmt3a in paternal and maternal imprinting. Nature 429(6994):900–903
Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y et al (2007) Role of the dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16(19):2272–2280
Kherraf Z, Cazin C, Bouker A, Mustapha FB, Hennebicq S, S. Septier A. et al (2022) Whole-exome sequencing improves the diagnosis and care of men with non-obstructive azoospermia. Am J Hum Genet 109(3):508–517
Lin X, Han M, Cheng L, Chen J, Zhang Z, Shen T et al (2016) Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. Rna Biol 13(10):1011–1024
Lin Z, Hsu P, Xing J, Fang X, Lu J, Z. Zou Q. et al (2017) Mettl3-/mettl14-mediated mRNA n(6)-methyladenosine modulates murine spermatogenesis. Cell Res 27(10):1216–1230
Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD (2008) Dicer1 is required for differentiation of the mouse male germline. Biol Reprod 79(4):696–703
Manfrevola F, Chioccarelli T, Cobellis G, Fasano S, Ferraro B, Sellitto C et al (2020) CircRNA role and circRNA-dependent network (ceRNET) in asthenozoospermia. Front Endocrinol 11:395
Misir S, Wu N, Yang B, B. (2022) Specific expression and functions of circular RNAs. Cell Death Differ 29(3):481–491
Monk M, Boubelik M, Lehnert S (1987) Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development (cambridge, England) 99(3):371–382
O’Donnell L, Nicholls P, K. O’Bryan M. K. McLachlan R. I. Stanton P. G. (2011) Spermiation: the process of sperm release. Spermatogenesis 1(1):14–35
Pang A, Rennert OM (2013) Protein acetylation and spermatogenesis. Reproductive System & Sexual Disorders S1:5
Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W (2002) Histone h2a variants h2ax and h2az. Curr Opin Genet Dev 12(2):162–169
Rogakou EP, Boon C, Redon C, Bonner WM (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146(5):905–916
Romero Y, Meikar O, Papaioannou M, Conne D, Grey B, C. Weier M. et al (2011) Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS ONE 6(10):e25241
Rotondo JC, Lanzillotti C, Mazziotta C, Tognon M, Martini F (2021) Epigenetics of male infertility: the role of DNA methylation. Frontiers in Cell and Developmental Biology 9:689624
Santos F, Dean W (2004) Epigenetic reprogramming during early development in mammals. Reproduction (cambridge, England) 127(6):643–651
Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y et al (2018) Alkbh5-dependent m6a demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. In Proceedings of the National Academy of Sciences 115(2):E325–E333
Testa E, Nardozi D, Antinozzi C, Faieta M, Di Cecca S, Caggiano C (2018) H2afx and mdc1 promote maintenance of genomic integrity in male germ cells. J Cell Sci, 131(6)
Wang S, Lv W, Li T, Zhang S, Wang H, Li X et al (2022) Dynamic regulation and functions of mRNA m6a modification. Cancer Cell Int 22(1):48
Xiang D, Zhu J, Hou C, Yang W (2014) Identification and expression pattern analysis of Piwi genes during the spermiogenesis of Portunus trituberculatus. Gene 534(2):240–248
Xu K, Yang YG, Feng H, Zhang XX, Wang C, Jiang L (2017a) Mettl3-mediated m6a regulates spermatogonial differentiation and meiosis initiation. Cell Res(27), 1100–1114
Xu K, Yang Y, Feng G, Sun B, Chen J, Li Y et al (2017b) Mettl3-mediated m(6)a regulates spermatogonial differentiation and meiosis initiation. Cell Res 27(9):1100–1114
Yu M, Mu H, Niu Z, Chu Z, Zhu H, Hua J (2014) Mir-34c enhances mouse spermatogonial stem cells differentiation by targeting nanos2. J Cell Biochem 115(2):232–242
Zhang Z, Wu H, Zheng L, Zhang H, Yang Y, Mao J (2022) Identification and characterization of circular RNAs in the testicular tissue of patients with non-obstructive azoospermia. Asian J Androl
Zhao J, Wang B, Yu H, Wang Y, Liu X, Zhang Q (2018) Tdrd1 is a germline-specific and sexually dimorphically expressed gene in Paralichthys olivaceus. Gene 673:61–69
Zheng G, Dahl J, Niu A, Fedorcsak Y, P. Huang C. M. Li C. J. et al (2013) Alkbh5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49(1):18–29
Zhou F, Chen W, Cui Y, Liu B, Yuan Q, Li Z et al (2020) Mirna-122-5p stimulates the proliferation and dna synthesis and inhibits the early apoptosis of human spermatogonial stem cells by targeting cbl and competing with lncrna casc7. Aging 12(24):25528–25546
Zhou F, Chen W, Jiang Y, He Z (2019) Regulation of long non-coding RNAs and circular RNAs in spermatogonial stem cells. Reproduction (cambridge, England) 158(1):R15–R25
Zhu Q, Kirby JA, Chu C. Gou L (2021) Small noncoding RNAs in reproduction and infertility. Biomedicines, 9(12)
Zimmermann C, Romero Y, Warnefors M, Bilican A, Borel C, Smith L, B. et al (2014) Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PLoS ONE 9(9):e107023
Acknowledgements
The authors would like to thank Dr Dev Sooranna of Imperial College in London for editing the manuscript.
Funding
This work was supported by grants from the Guangxi Natural Science Foundation (No. 2020GXNSFBA297148 and AD20159062), the National Natural Science Foundation of China (No. 32102514, 31972996 and U20A2051), the Guangxi Science and Technology Major Project (No. GuiKe AA22068099), and the Bama County Program for Talents in Science and Technology, Guangxi, China (No. 202101265 and 202100174).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Informed consent
Dandan Zhong, Liyin Zhang, and Kongwei Huang were responsible for the execution of the experiments and drawing diagrams. Dandan Zhong and Kongwei Huang participated in the writing of the manuscript. Kongwei Huang analyzed the sequencing data. Mengjie Chen and Yaling Chen were responsible for the analysis of experimental data. Qingyou Liu and Deshun Shi were involved in the revision of the manuscript. Hui Li was responsible for the experimental design and guided the project. Dandan Zhong and Liyin Zhang contributed equally to this work. All the authors read and approved the final manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhong, D., Zhang, L., Huang, K. et al. circRNA-miRNA-mRNA network analysis to explore the pathogenesis of abnormal spermatogenesis due to aberrant m6A methylation. Cell Tissue Res 392, 605–620 (2023). https://doi.org/10.1007/s00441-022-03725-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-022-03725-7