Abstract
Purpose
Due to the rarity of primary gastrointestinal lymphoma (PGIL), the prognostic factors and optimal management of PGIL have not been clearly defined. We aimed to establish prognostic models using a deep learning algorithm for survival prediction.
Methods
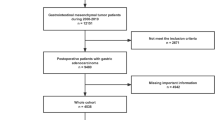
We collected 11,168 PGIL patients from the Surveillance, Epidemiology, and End Results (SEER) database to form the training and test cohorts. At the same time, we collected 82 PGIL patients from three medical centres to form the external validation cohort. We constructed a Cox proportional hazards (CoxPH) model, random survival forest (RSF) model, and neural multitask logistic regression (DeepSurv) model to predict PGIL patients’ overall survival (OS).
Results
The 1-, 3-, 5-, and 10-year OS rates of PGIL patients in the SEER database were 77.1%, 69.4%, 63.7%, and 50.3%, respectively. The RSF model based on all variables showed that the top three most important variables for predicting OS were age, histological type, and chemotherapy. The independent risk factors for PGIL patient prognosis included sex, age, race, primary site, Ann Arbor stage, histological type, symptom, radiotherapy, and chemotherapy, according to the Lasso regression analysis. Using these factors, we built the CoxPH and DeepSurv models. The DeepSurv model’s C-index values were 0.760 in the training cohort, 0.742 in the test cohort, and 0.707 in the external validation cohort, which demonstrated that the DeepSurv model performed better compared to the RSF model (0.728) and the CoxPH model (0.724). The DeepSurv model accurately predicted 1-, 3-, 5- and 10-year OS. Both calibration curves and decision curve analysis curves demonstrated the superior performance of the DeepSurv model. We developed the DeepSurv model as an online web calculator for survival prediction, which can be accessed at http://124.222.228.112:8501/.
Conclusions
This DeepSurv model with external validation is superior to previous studies in predicting short-term and long-term survival and can help us make better-individualized decisions for PGIL patients.







Similar content being viewed by others
Data availability
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
References
Adham D, Abbasgholizadeh N, Abazari M (2017) Prognostic factors for survival in patients with gastric cancer using a random survival forest. Asian Pac J Cancer Prev 18(1):129–134. https://doi.org/10.22034/APJCP.2017.18.1.129
Alvarez-Lesmes J, Chapman JR, Cassidy D, Zhou Y, Garcia-Buitrago M, Montgomery EA, Lossos IS, Sussman D, Poveda J (2021) Gastrointestinal tract lymphomas. Arch Pathol Lab Med 145(12):1585–1596. https://doi.org/10.5858/arpa.2020-0661-RA
Bautista-Quach MA, Ake CD, Chen M, Wang J (2012) Gastrointestinal lymphomas: morphology, immunophenotype and molecular features. J Gastrointest Oncol 3(3):209–225. https://doi.org/10.3978/j.issn.2078-6891.2012.024
Cheung MC, Housri N, Ogilvie MP, Sola JE, Koniaris LG (2009) Surgery does not adversely affect survival in primary gastrointestinal lymphoma. J Surg Oncol 100(1):59–64. https://doi.org/10.1002/jso.21298
Choi KH, Lee HH, Jung SE, Park KS, Joo-Hyun O, Jeon YW, Choi BO, Cho SG (2020) Analysis of the response time to involved-field radiotherapy in primary gastrointestinal low-grade B-cell lymphoma. Radiat Oncol 15(1):210. https://doi.org/10.1186/s13014-020-01649-6
Chunli Y, Ming J, Ziyan M, Jie J, Shuli L, Jie H, Yu W, Caigang X, Liqun Z (2023) Real-world clinical features and survival outcomes associated with primary gastrointestinal natural killer/T-cell lymphoma from 1999 to 2020. Cancer Med 12(3):2614–2623. https://doi.org/10.1002/cam4.5136
Ghimire P, Wu GY, Zhu L (2011) Primary gastrointestinal lymphoma. World J Gastroenterol 17(6):697–707. https://doi.org/10.3748/wjg.v17.i6.697
Goerdten J, Carrière I, Muniz-Terrera G (2020) Comparison of Cox proportional hazards regression and generalized Cox regression models applied in dementia risk prediction. Alzheimers Dement (n y) 6(1):e12041. https://doi.org/10.1002/trc2.12041
Hadanny A, Shouval R, Wu J, Gale CP, Unger R, Zahger D, Gottlieb S, Matetzky S, Goldenberg I, Beigel R, Iakobishvili Z (2022) Machine learning-based prediction of 1-year mortality for acute coronary syndrome. J Cardiol 79(3):342–351. https://doi.org/10.1016/j.jjcc.2021.11.006
Huang J, Jiang W, Xu R, Huang H, Lv Y, Xia Z, Sun X, Guan Z, Lin T, Li Z (2010) Primary gastric non-Hodgkin’s lymphoma in chinese patients: clinical characteristics and prognostic factors. BMC Cancer 10:358. https://doi.org/10.1186/1471-2407-10-358
Ivanics T, Nelson W, Patel MS, Claasen MPAW, Lau L, Gorgen A, Abreu P, Goldenberg A, Erdman L, Sapisochin G (2022) The Toronto postliver transplantation hepatocellular carcinoma recurrence calculator: a machine learning approach. Liver Transpl 28(4):593–602. https://doi.org/10.1002/lt.26332
Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y (2018) DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol 18(1):24. https://doi.org/10.1186/s12874-018-0482-1
Kohri M, Tsukasaki K, Akuzawa Y, Tanae K, Takahashi N, Saeki T, Okamura D, Ishikawa M, Maeda T, Kawai N, Matsuda A, Arai E, Arai S, Asou N (2020) Peripheral T-cell lymphoma with gastrointestinal involvement and indolent T-lymphoproliferative disorders of the gastrointestinal tract. Leuk Res 91:106336. https://doi.org/10.1016/j.leukres.2020.106336
Kühnl A, Cunningham D, Counsell N, Hawkes EA, Qian W, Smith P, Chadwick N, Lawrie A, Mouncey P, Jack A, Pocock C, Ardeshna KM, Radford J, McMillan A, Davies J, Turner D, Kruger A, Johnson PW, Gambell J, Rosenwald A, Ott G, Horn H, Ziepert M, Pfreundschuh M, Linch D (2017) Outcome of elderly patients with diffuse large B-cell lymphoma treated with R-CHOP: results from the UK NCRI R-CHOP14v21 trial with combined analysis of molecular characteristics with the DSHNHL RICOVER-60 trial. Ann Oncol 28(7):1540–1546. https://doi.org/10.1093/annonc/mdx128
Lu Y, Liu H, Zhuo X, Liu T, Lou X, Li C, Zhi M (2023) Clinical, endoscopic and pathological characteristics of primary gastrointestinal T-cell and NK/T-cell lymphomas. Scand J Gastroenterol 58(2):163–169. https://doi.org/10.1080/00365521.2022.2114811
Mehmet K, Sener C, Uyeturk U, Seker M, Tastekin D, Tonyali O, Balakan O, Yazici OK, Urakci Z, Isikdogan A, Ozdemir N, Inal A, Kaplan MA, Suner A, Dal S, Uncu D, Gumus M, Boruban MC, Oksuzoglu B, Ayyildiz O, Benekli M (2014) Treatment modalities in primary gastric lymphoma: the effect of rituximab and surgical treatment. A study by the Anatolian Society of Medical Oncology. Contemp Oncol (pozn) 18(4):273–278. https://doi.org/10.5114/wo.2014.40556
Nakamura S, Matsumoto T, Iida M, Yao T, Tsuneyoshi M (2003) Primary gastrointestinal lymphoma in Japan: a clinicopathologic analysis of 455 patients with special reference to its time trends. Cancer 97(10):2462–2473. https://doi.org/10.1002/cncr.11415
Olszewska-Szopa M, Wróbel T (2019) Gastrointestinal non-Hodgkin lymphomas. Adv Clin Exp Med 28(8):1119–1124. https://doi.org/10.17219/acem/94068
Ouyang D, Shi M, Wang Y, Luo L, Huang L (2022) Prognostic analysis of pT1-T2aN0M0 cervical adenocarcinoma based on random survival forest analysis and the generation of a predictive nomogram. Front Oncol 12:1049097. https://doi.org/10.3389/fonc.2022.1049097
Peng JC, Zhong L, Ran ZH (2015) Primary lymphomas in the gastrointestinal tract. J Dig Dis 16(4):169–176. https://doi.org/10.1111/1751-2980.12234
Randall RL, Cable MG (2016) Nominal nomograms and marginal margins: what is the law of the line? Lancet Oncol 17(5):554–556. https://doi.org/10.1016/S1470-2045(16)00072-3
Shen Y, Ou J, Wang B, Wang L, Xu J, Cen X (2021) Influence of severe gastrointestinal complications in primary gastrointestinal diffuse large B-cell lymphoma. Cancer Manag Res 13:1041–1052. https://doi.org/10.2147/CMAR.S295671
Sun B, Xia Y, Guo Y, He C, Wang W (2020) Development and validation of prognostic nomograms for patients with primary gastrointestinal non-Hodgkin lymphomas. Dig Dis Sci 65(12):3570–3582. https://doi.org/10.1007/s10620-020-06078-9
Taylor JM (2011) Random survival forests. J Thorac Oncol 6(12):1974–1975. https://doi.org/10.1097/JTO.0b013e318233d835
Wang J, Zhou M, Zhou R, Xu J, Chen B (2020) Nomogram for predicting the overall survival of adult patients with primary gastrointestinal diffuse large b cell lymphoma: a SEER-based study. Front Oncol 10:1093. https://doi.org/10.3389/fonc.2020.01093
Wang J, Ren W, Zhang C, Wang X (2022) A new staging system based on the dynamic prognostic nomogram for elderly patients with primary gastrointestinal diffuse large B-cell lymphoma. Front Med (lausanne) 9:860993. https://doi.org/10.3389/fmed.2022.860993
Wu WT, Li YJ, Feng AZ, Li L, Huang T, Xu AD, Lyu J (2021) Data mining in clinical big data: the frequently used databases, steps, and methodological models. Mil Med Res 8(1):44. https://doi.org/10.1186/s40779-021-00338-z
Xie Y, Jia M, Shi J, Tao Y (2020) Inferior prognosis of gastric involvement in patients with gastrointestinal burkitt lymphoma. Cancer Med 9(9):3107–3114. https://doi.org/10.1002/cam4.2975
Yang L, Tang X, Peng X, Qian D, Guo Q, Guo H (2018) Clinical characteristics of primary intestinal NK/T cell lymphoma, nasal type: case series and review of the literature. Pathol Res Pract 214(8):1081–1086. https://doi.org/10.1016/j.prp.2018.05.013
Yang J, Li Y, Liu Q, Li L, Feng A, Wang T, Zheng S, Xu A, Lyu J (2020) Brief introduction of medical database and data mining technology in big data era. J Evid Based Med 13(1):57–69. https://doi.org/10.1111/jebm.12373
Zhou H, Huang Y, Qiu Z, Zhao H, Fang W, Yang Y, Zhao Y, Hou X, Ma Y, Hong S, Zhou T, Zhang Y, Zhang L (2018) Impact of Prior cancer history on the overall survival of patients newly diagnosed with cancer: a pan-cancer analysis of the SEER database. Int J Cancer 143(7):1569–1577. https://doi.org/10.1002/ijc.31543
Acknowledgments
The authors thank all the medical researchers and staff who participated in maintaining the SEER database.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
ZXL and LHL: Conceptualization. FFW and LC: methodology. FFW: software. LHL, YTJ, and WL: validation. WL, LJW, JZ, WL, and WJL: data curation. FFW: writing—original draft preparation. ZXL: writing—review and editing. LC: visualization. ZXL: supervision. ZXL: project administration. ZXL: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the First Hospital of Hebei Medical University (10 May 2023, 2023198).
Informed consent
Patient consent was waived due to the retrospective nature of the study. All data used in this manuscript were anonymized.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, F., Chen, L., Liu, L. et al. Deep learning model for predicting the survival of patients with primary gastrointestinal lymphoma based on the SEER database and a multicentre external validation cohort. J Cancer Res Clin Oncol 149, 12177–12189 (2023). https://doi.org/10.1007/s00432-023-05123-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05123-0