Abstract
Main conclusion
Gene co-expression network analysis of the heat-responsive core transcriptome in two contrasting Brassica rapa accessions reveals the main metabolic pathways, key modules and hub genes, are involved in long-term heat stress.
Abstract
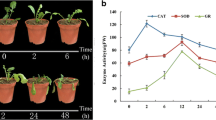
Brassica rapa is a widely cultivated and economically important vegetable in Asia. High temperature is a common stress that severely impacts leaf head formation in B. rapa, resulting in reduced quality and production. The purpose of this study was thus to identify candidate heat tolerance genes by comparative transcriptome analysis of two contrasting B. rapa accessions in response to long-term heat stress. Two B. rapa accessions, ‘268’ and ‘334’, which showed significant differences in heat tolerance, were used for RNA sequencing analysis. We identified a total of 11,055 and 8921 differentially expressed genes (DEGs) in ‘268’ and ‘334’, respectively. Functional enrichment analyses of all of the identified DEGs, together with the genes identified from weighted gene co‐expression network analyses (WGCNA), revealed that the autophagy pathway, glutathione metabolism, and ribosome biogenesis in eukaryotes were significantly up-regulated, whereas photosynthesis was down-regulated, in the heat resistance of B. rapa ‘268’. Furthermore, when B. rapa ‘334’ was subjected to long-term high-temperature stress, heat stress caused significant changes in the expression of certain functional genes linked to protein processing in the endoplasmic reticulum and plant hormone signal transduction pathways. Autophagy-related genes might have been induced by persistent heat stress and remained high during recovery. Several hub genes like HSP17.6, HSP17.6B, HSP70-8, CLPB1, PAP1, PYR1, ADC2, and GSTF11 were discussed in this study, which may be potential candidates for further analyses of the response to long-term heat stress. These results should help elucidate the molecular mechanisms of heat stress adaptation in B. rapa.










Similar content being viewed by others
Abbreviations
- CK:
-
Control treatment
- DEGs:
-
Differentially expressed genes
- FPKM:
-
Fragments per kilo-base per million reads
- GO:
-
Gene ontology enrichment
- HSF:
-
Heat shock transcription factor
- HSP:
-
Heat shock protein
- HT:
-
Heat stress treatment
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- RC:
-
Recovery treatment
- WGCNA:
-
Weighted gene co-expression network analysis
References
Amrine KC, Blanco-Ulate B, Cantu D (2015) Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis. PLoS ONE 10:e0118731. https://doi.org/10.1371/journal.pone.0118731
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. https://doi.org/10.1186/gb-2010-11-10-r106
Ashraf MF, Yang S, Wu R, Wang Y, Hussain A, Noman A, Khan MI, Liu Z, Qiu A, Guan D, He S (2018) Capsicum annuum HsfB2a positively regulates the response to Ralstonia solanacearum infection or high temperature and high humidity forming transcriptional cascade with CaWRKY6 and CaWRKY40. Plant Cell Physiol 59:2608–2623. https://doi.org/10.1093/pcp/pcy181
Avelange-Macherel MH, Payet N, Lalanne D, Neveu M, Tolleter D, Burstin J, Macherel D (2015) Variability within a pea core collection of LEAM and HSP22, two mitochondrial seed proteins involved in stress tolerance. Plant Cell Environ 38:1299–1311. https://doi.org/10.1111/pce.12480
Avin-Wittenberg T (2019) Autophagy and its role in plant abiotic stress management. Plant Cell Environ 42:1045–1053. https://doi.org/10.1111/pce.13404
Awasthi R, Bhandari K, Nayyar H (2015) Temperature stress and redox homeostasis in agricultural crops. Front Environ Sci 3:11. https://doi.org/10.3389/fenvs.2015.00011
Bari R, Jones JDG (2008) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488. https://doi.org/10.1007/s11103-008-9435-0
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. https://doi.org/10.2307/2346101
Bokszczanin KL, Network SPTIT, C, Fragkostefanakis S, (2013) Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front Plant Sci 4:315. https://doi.org/10.3389/fpls.2013.00315
Challinor AJ, Watson J, Lobell DB, Howden SM, Smith DR, Chhetri N (2014) A meta-analysis of crop yield under climate change and adaptation. Nat Clim Chang 4:287–291. https://doi.org/10.1038/nclimate2153
Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X (2011) BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11:136. https://doi.org/10.1186/1471-2229-11-136
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8(Suppl 4):S11. https://doi.org/10.1186/1752-0509-8-S4-S11
Dai Y, Zhang S, Sun X, Li G, Yuan L, Li F, Zhang H, Zhang S, Chen G, Wang C, Sun R (2020) Comparative transcriptome analysis of gene expression and regulatory characteristics associated with different vernalization periods in Brassica rapa. Genes (Basel). https://doi.org/10.3390/genes11040392
DeRocher AE, Helm KW, Lauzon LM, Vierling E (1991) Expression of a conserved family of cytoplasmic low molecular weight heat shock proteins during heat stress and recovery. Plant Physiol 96:1038–1047. https://doi.org/10.1104/pp.96.4.1038
Dong X, Yi H, Lee J, Nou IS, Han CT, Hur Y (2015) Global gene-expression analysis to identify differentially expressed genes critical for the heat stress response in Brassica rapa. PLoS ONE 10:e0130451. https://doi.org/10.1371/journal.pone.0130451
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95(25):14863–14868. https://doi.org/10.1073/pnas.95.25.14863
Guo M, Liu JH, Ma X, Zhai YF, Gong ZH, Lu MH (2016) Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70–2 involvement in heat stress. Plant Sci 252:246–256. https://doi.org/10.1016/j.plantsci.2016.07.001
Han Y, Yong X, Yu J, Cheng T, Wang J, Yang W, Pan H, Zhang Q (2019) Identification of candidate adaxial-abaxial-related genes regulating petal expansion during flower opening in Rosa chinensis “Old Blush.” Front Plant Sci 10:1098. https://doi.org/10.3389/fpls.2019.01098
Hong S-W, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97:4392–4397. https://doi.org/10.1073/pnas.97.8.4392
Hopper DW, Ghan R, Schlauch KA, Cramer GR (2016) Transcriptomic network analyses of leaf dehydration responses identify highly connected ABA and ethylene signaling hubs in three grapevine species differing in drought tolerance. BMC Plant Biol 16:118. https://doi.org/10.1186/s12870-016-0804-6
Hu W, Hu G, Han B (2009) Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci 176:583–590. https://doi.org/10.1016/j.plantsci.2009.01.016
Huang XY, Tao P, Li BY, Wang WH, Yue ZC, Lei JL, Zhong XM (2015) Genome-wide identification, classification, and analysis of heat shock transcription factor family in Chinese cabbage (Brassica rapa pekinensis). Genet Mol Res 14:2189–2204. https://doi.org/10.4238/2015.March.27.5
Isah T (2019) Stress and defense responses in plant secondary metabolites production. Biol Res 52:39. https://doi.org/10.1186/s40659-019-0246-3
Jacob P, Hirt H, Bendahmane A (2017) The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol J 15:405–414. https://doi.org/10.1111/pbi.12659
Julius BT, Leach KA, Tran TM, Mertz RA, Braun DM (2017) Sugar transporters in plants: new insights and discoveries. Plant Cell Physiol 58:1442–1460. https://doi.org/10.1093/pcp/pcx090
Kaushal N, Awasthi R, Gupta K, Gaur P, Siddique KHM, Nayyar H (2013) Heat-stress-induced reproductive failures in chickpea (Cicer arietinum) are associated with impaired sucrose metabolism in leaves and anthers. Funct Plant Biol 40:1334–1349. https://doi.org/10.1071/FP13082
Kost MA, Perales HR, Wijeratne S, Wijeratne AJ, Stockinger E, Mercer KL (2017) Differentiated transcriptional signatures in the maize landraces of Chiapas, Mexico. BMC Genom 18:707. https://doi.org/10.1186/s12864-017-4005-y
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. https://doi.org/10.1186/1471-2105-9-559
Lee U, Rioflorido I, Hong SW, Larkindale J, Waters ER, Vierling E (2007) The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J 49:115–127. https://doi.org/10.1111/j.1365-313X.2006.02940.x
Lee SS, Jung WY, Park HJ, Lee A, Kwon SY, Kim HS, Cho HS (2018) Genome-wide analysis of alternative splicing in an inbred Cabbage (Brassica oleracea L.) line “HO” in response to heat stress. Curr Genom 19:12–20. https://doi.org/10.2174/1389202918666170705151901
Li H, Liu SS, Yi CY, Wang F, Zhou J, Xia XJ, Shi K, Zhou YH, Yu JQ (2014) Hydrogen peroxide mediates abscisic acid-induced HSP70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ 37:2768–2780. https://doi.org/10.1111/pce.12360
Li S, Yu J, Li Y, Zhang H, Bao X, Bian J, Xu C, Wang X, Cai X, Wang Q, Wang P, Guo S, Miao Y, Chen S, Qin Z, Dai S (2019) Heat-responsive proteomics of a heat-sensitive Spinach variety. Int J Mol Sci 20:3872. https://doi.org/10.3390/ijms20163872
Lin CW, Fu SF, Liu YJ, Chen CC, Chang CH, Yang YW, Huang HJ (2019) Analysis of ambient temperature-responsive transcriptome in shoot apical meristem of heat-tolerant and heat-sensitive broccoli inbred lines during floral head formation. BMC Plant Biol 19:3. https://doi.org/10.1186/s12870-018-1613-x
Liu J, Feng L, Li J, He Z (2015) Genetic and epigenetic control of plant heat responses. Front Plant Sci 6:267. https://doi.org/10.3389/fpls.2015.00267
Liu J, Pang X, Cheng Y, Yin Y, Zhang Q, Su W, Hu B, Guo Q, Ha S, Zhang J, Wan H (2018) The Hsp70 gene family in Solanum tuberosum: genome-wide identification, phylogeny, and expression patterns. Sci Rep 8:16628. https://doi.org/10.1038/s41598-018-34878-7
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Luo Y, Pang D, Jin M, Chen J, Kong X, Li W, Chang Y, Li Y, Wang Z (2019) Identification of plant hormones and candidate hub genes regulating flag leaf senescence in wheat response to water deficit stress at the grain-filling stage. Plant Direct 3:e00152. https://doi.org/10.1002/pld3.152
Merlot S, Gost F, Dl G, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25:295–303. https://doi.org/10.1046/j.1365-313x.2001.00965.x
Miller G, Shulaev V, Mittler R (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133:481–489. https://doi.org/10.1111/j.1399-3054.2008.01090.x
Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105:9433–9438. https://doi.org/10.1073/pnas.0803611105
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185. https://doi.org/10.1146/annurev.arplant.56.032604.144046
Nieto-Sotelo J, Martínez LM, Ponce G, Cassab GI, Alagón A, Meeley RB, Ribaut JM, Yang R (2002) Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. Plant Cell 14:1621–1633. https://doi.org/10.1105/tpc.010487
Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G (2009) Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21:403–419. https://doi.org/10.1105/tpc.108.064691
Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22:53–65. https://doi.org/10.1016/j.tplants.2016.08.015
Park E, Kim T-H (2017) Production of ABA responses requires both the nuclear and cytoplasmic functional involvement of PYR1 Biochemical and Biophysical. Res Commun 484:34–39. https://doi.org/10.1016/j.bbrc.2017.01.082
Perez-Amador MA (2002) Induction of the arginine decarboxylase ADC2 gene provides evidence for the involvement of polyamines in the wound response in Arabidopsis. Plant Physiol 130:1454–1463. https://doi.org/10.1104/pp.009951
Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G (2013) Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol 23:310–322. https://doi.org/10.1016/j.semcancer.2013.05.008
Prasad BD, Goel S, Krishna P (2010) In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS ONE 5:e12761. https://doi.org/10.1371/journal.pone.0012761
Qi C, Lin X, Li S, Liu L, Wang Z, Li Y, Bai R, Xie Q, Zhang N, Ren S, Zhao B, Li X, Fan S, Guo YD (2019) SoHSC70 positively regulates thermotolerance by alleviating cell membrane damage, reducing ROS accumulation, and improving activities of antioxidant enzymes. Plant Sci 283:385–395. https://doi.org/10.1016/j.plantsci.2019.03.003
Qian Y, Cao L, Zhang Q, Amee M, Chen K, Chen L (2020) SMRT and Illumina RNA sequencing reveal novel insights into the heat stress response and crosstalk with leaf senescence in tall fescue. BMC Plant Biol 20:366. https://doi.org/10.1186/s12870-020-02572-4
Qu AL, Ding YF, Jiang Q, Zhu C (2013) Molecular mechanisms of the plant heat stress response Biochem Biophys. Res Commun 432:203–207. https://doi.org/10.1016/j.bbrc.2013.01.104
Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12:479–492. https://doi.org/10.1105/tpc.12.4.479
Ray DK, Gerber JS, MacDonald GK, West PC (2015) Climate variation explains a third of global crop yield variability. Nat Commun 6:5989. https://doi.org/10.1038/ncomms6989
Reinbothe C, Springer A, Samol I, Reinbothe S (2009) Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J 276:4666–4681. https://doi.org/10.1111/j.1742-4658.2009.07193.x
Sagor GHM, Berberich T, Takahashi Y, Niitsu M, Kusano T (2012) The polyamine spermine protects Arabidopsis from heat stress-induced damage by increasing expression of heat shock-related genes. Transgen Res 22:595–605. https://doi.org/10.1007/s11248-012-9666-3
Saleem MA, Malik W, Qayyum A, Ul-Allah S, Ahmad MQ, Afzal H, Amjid MW, Ateeq MF, Zia ZU (2021) Impact of heat stress responsive factors on growth and physiology of cotton (Gossypium hirsutum L.). Mol Biol Rep 48:1069–1079. https://doi.org/10.1007/s11033-021-06217-z
Sato S, Kato N, Iwai S, Hagimori M (2002) Effect of low temperature pretreatment of buds or inflorescence on isolated microspore culture in Brassica rapa (syn. B. campestris). Breeding Sci 52:23–26. https://doi.org/10.1270/jsbbs.52.23
Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819:104–119. https://doi.org/10.1016/j.bbagrm.2011.10.002
Sedaghatmehr M, Thirumalaikumar VP, Kamranfar I, Marmagne A, Masclaux-Daubresse C, Balazadeh S (2019) A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ 42:1054–1064. https://doi.org/10.1111/pce.13426
Shan X, Wang J, Chua L, Jiang D, Peng W, Xie D (2011) The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol 155:751–764. https://doi.org/10.1104/pp.110.166595
Singh D, Singh CK, Taunk J, Jadon V, Pal M, Gaikwad K (2019) Genome wide transcriptome analysis reveals vital role of heat responsive genes in regulatory mechanisms of lentil (Lens culinaris Medikus). Sci Rep 9:12976. https://doi.org/10.1038/s41598-019-49496-0
Song X, Liu G, Duan W, Liu T, Huang Z, Ren J, Li Y, Hou X (2014) Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol Genet Genom 289:541–551. https://doi.org/10.1007/s00438-014-0833-5
Takehisa H, Sato Y (2019) Transcriptome monitoring visualizes growth stage-dependent nutrient status dynamics in rice under field conditions. Plant J 97:1048–1060. https://doi.org/10.1111/tpj.14176
Tan M, Cheng D, Yang Y, Zhang G, Qin M, Chen J, Chen Y, Jiang M (2017) Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol 17:194. https://doi.org/10.1186/s12870-017-1143-y
Thatcher LF, Foley R, Casarotto HJ, Gao LL, Kamphuis LG, Melser S, Singh KB (2018) The RNA polymerase II carboxyl terminal domain (CTD) phosphatase-Like1 (CPL1) is a biotic stress susceptibility gene. Sci Rep 8:13454. https://doi.org/10.1038/s41598-018-31837-0
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. https://doi.org/10.1093/bioinformatics/btp120
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. https://doi.org/10.1038/nprot.2012.016
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86. https://doi.org/10.1186/s12870-016-0771-y
Wang Y, Lin S, Song Q, Li K, Tao H, Huang J, Chen X, Que S, He H (2014) Genome-wide identification of heat shock proteins (Hsps) and Hsp interactors in rice: Hsp70s as a case study. BMC Genom 15:344. https://doi.org/10.1186/1471-2164-15-344
Wang A, Hu J, Huang X, Li X, Zhou G, Yan Z (2016) Comparative transcriptome analysis reveals heat-responsive genes in Chinese cabbage (Brassica rapa ssp. chinensis). Front Plant Sci 7:939. https://doi.org/10.3389/fpls.2016.00939
Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100:681–697. https://doi.org/10.1093/aob/mcm079
Wijewardene I, Mishra N, Sun L, Smith J, Zhu X, Payton P, Shen G, Zhang H (2020) Improving drought-, salinity-, and heat-tolerance in transgenic plants by co-overexpressing Arabidopsis vacuolar pyrophosphatase gene AVP1 and Larrea Rubisco activase gene RCA. Plant Sci 296:110499. https://doi.org/10.1016/j.plantsci.2020.110499
Wu Y, Xu J, Han X, Qiao G, Yang K, Wen Z, Wen X (2020) Comparative transcriptome analysis combining SMRT- and Illumina-based RNA-Seq identifies potential candidate genes involved in betalain biosynthesis in Pitaya fruit. Int J Mol Sci 21:3288. https://doi.org/10.3390/ijms21093288
Xie L, Dong C, Shang Q (2019) Gene co-expression network analysis reveals pathways associated with graft healing by asymmetric profiling in tomato. BMC Plant Biol 19:373. https://doi.org/10.1186/s12870-019-1976-7
Xu J, Xue C, Xue D, Zhao J, Gai J, Guo N, Xing H (2013) Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE 8:e69810. https://doi.org/10.1371/journal.pone.0069810
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, Prins M, van Tunen A, He Y (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot 63:1025–1038. https://doi.org/10.1093/jxb/err337
Zhang L, Cai X, Wu J, Liu M, Grob S, Cheng F, Liang J, Cai C, Liu Z, Liu B, Wang F, Li S, Liu F, Li X, Cheng L, Yang W, Li MH, Grossniklaus U, Zheng H, Wang X (2018a) Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res 5:50. https://doi.org/10.1038/s41438-018-0071-9
Zhang S, Wang S, Lv J, Liu Z, Wang Y, Ma N, Meng Q (2018b) SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato. Plant Cell Physiol 59:58–71. https://doi.org/10.1093/pcp/pcx160
Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z (2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 9:e1003196. https://doi.org/10.1371/journal.pgen.1003196
Zhou J, Wang J, Yu JQ, Chen Z (2014) Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci 5:174. https://doi.org/10.3389/fpls.2014.00174
Acknowledgements
We thank Prof. Cheng Feng and Dr. Zhang Kang (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China) for help and advice on transcriptome data analysis. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (Grant number 2017YFD0101802) and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS). This work was performed at the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing, China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yue, L., Li, G., Dai, Y. et al. Gene co-expression network analysis of the heat-responsive core transcriptome identifies hub genes in Brassica rapa. Planta 253, 111 (2021). https://doi.org/10.1007/s00425-021-03630-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03630-3