Abstract
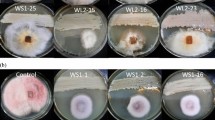
The identification of an increasing number of drug-resistant pathogens has stimulated the development of new therapeutic agents to combat them. Microbial natural products are among the most important elements when it comes to drug discovery. Today, thiopeptide antibiotics are receiving increasing research attention due to their potent activity against Gram-positive bacteria. In this study, we demonstrated the successful use of a whole-cell microbial biosensor (Streptomyces lividans TK24 pMO16) for the specific detection of thiopeptide antibiotics among the native actinomycete strains isolated from the rhizosphere soil of Juniperus excelsa (Bieb.). Among the native strains, two strains of Streptomyces, namely sp. Je 1–79 and Je 1–613, were identified that were capable of producing thiopeptide antibiotics. A multilocus sequence analysis of five housekeeping genes (gyrB, atpD, recA, rpoB, and trpB) classified them as representatives of two different species of the genus Streptomyces. The thiopeptide antibiotics berninamycin A and B were identified in the extracts of the two strains by means of a dereplication analysis. The berninamycin biosynthetic gene cluster was also detected in the genome of the Streptomyces sp. Je 1–79 strain and showed a high level of similarity (93%) with the ber cluster from S. bernensis. Thus, the use of this whole-cell biosensor during the first stage of the screening process could serve to accelerate the specific detection of thiopeptide antibiotics.





Similar content being viewed by others
Data Availability
Not applicable.
Code Availability
Not applicable.
References
Ventola CL (2015) The antibiotic resistance crisis: part 1: causes and threats. PT 40(4):277–283
Mast Y, Stegmann E (2019) Actinomycetes: the antibiotics producers. Antibiotics (Basel) 8(3):105. https://doi.org/10.3390/antibiotics8030105
Genilloud O (2017) Actinomycetes: still a source of novel antibiotics. Nat Prod Rep 34:1203–1232. https://doi.org/10.1039/c7np00026j
Balouiri M, Sadiki M, Ibnsouda SK (2016) Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal 6(2):71–79. https://doi.org/10.1016/j.jpha.2015.11.005
Park M, Tsai SL, Chen W (2013) Microbial biosensors: engineered microorganisms as the sensing machinery. Sensors (Basel) 13(5):5777–5795. https://doi.org/10.3390/s130505777
Mascher T, Zimmer SL, Smith T-A, Helmann JD (2004) Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48(8):2888–2896. https://doi.org/10.1128/AAC.48.8.2888-2896.2004
Revilla-Guarinos A, Dürr F, Popp PF et al (2020) Amphotericin B specifically induces the two-component system LnrJK: development of a novel whole-cell biosensor for the detection of amphotericin-like polyenes. Front Microbiol 11:2022. https://doi.org/10.3389/fmicb.2020.02022
Rebets Y, Schmelz S, Gromyko O et al (2018) Design, development and application of whole-cell based antibiotic-specific biosensor. Metab Eng 47:263–270. https://doi.org/10.1016/j.ymben.2018.03.019
Bagley MC, Dale JW, Merritt EA, Xiong X (2005) Thiopeptide antibiotics. Chem Rev 105(2):685–714. https://doi.org/10.1021/cr0300441
Su TL (1948) Micrococcin, an antibacterial substance formed by a strain of Micrococcus. Br J Exp Pathol 29(5):473–481
Li J, Qu X, He X et al (2012) ThioFinder: a webbased tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS ONE 7:e45878. https://doi.org/10.1371/journal.pone.0045878
Wang W, Park K-H, Lee J et al (2020) A new thiopeptide antibiotic, Micrococcin P3, from a marine-derived strain of the bacterium Bacillus stratosphericus. Molecules 25(19):4383. https://doi.org/10.3390/molecules25194383
Iniyan AM, Sudarman E, Wink J et al (2019) Ala-geninthiocin, a new broad spectrum thiopeptide antibiotic, produced by a marine Streptomyces sp. ICN19. J Antibiot 72(2):99–105. https://doi.org/10.1038/s41429-018-0115-2
Shen X, Mustafa M, Chen Y et al (2019) Natural thiopeptides as a privileged scaffold for drug discovery and therapeutic development. Med Chem Res 28(8):1063–1098. https://doi.org/10.1007/s00044-019-02361-1
Ranieri M, Chan D, Yaeger LN et al (2019) Thiostrepton hijacks pyoverdine receptors to inhibit growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e00472-e519. https://doi.org/10.1128/AAC.00472-19
Degiacomi G, Personne Y, Mondesert G et al (2016) Micrococcin P1—a bactericidal thiopeptide active against Mycobacterium tuberculosis. Tuberculosis (Edinb) 100:95–101. https://doi.org/10.1016/j.tube.2016.07.011
Béahdy J (1974) Recent developments of antibiotic research and classification of antibiotics according to chemical structure. Adv Appl Microbiol 18:309–406
Hughes RA, Moody CJ (2007) From amino acids to heteroaromatics—thiopeptide antibiotics, nature’s heterocyclic peptides. Angew Chem Int Ed Engl 46(42):7930–7954. https://doi.org/10.1002/anie.200700728
Raju R, Gromyko O, Fedorenko V et al (2012) Juniperolide A: a new polyketide isolated from a terrestrial actinomycete Streptomyces sp. Org Lett 14(23):5860–5863. https://doi.org/10.1021/ol302766z
Raju R, Gromyko O, Fedorenko V et al (2012) Leopolic acid A, isolated from a terrestrial actinomycete, Streptomyces sp. Tetrahedron Lett 53:6300–6301. https://doi.org/10.1016/j.tetlet.2012.09.046
Tistechok SI, Tymchuk IV, Korniychuk OP et al (2021) Genetic identification and antimicrobial activity of Streptomyces sp. strain Je 1–6 isolated from Rhizosphere soil of Juniperus excelsa Bieb. Cytol Genet 55:28–35. https://doi.org/10.3103/S0095452721010138
Tistechok SI, Syrvatka VJ, Fedorenko VO, Gromyko OM (2018) Actinomycetes of Juniperus excelsa Bield. Rhizosphere: antagonists of phytopathogenic microbiota. Faktori eksperimental’noi evolucii organizmiv 23:340–345 ((in Ukrainian))
Myronovskyy M, Ostash B, Ostash I, Fedorenko V (2009) A gene cloning system for the siomycin producer Streptomyces sioyaensis NRRL-B5408. Folia Microbiol 54(2):91–96. https://doi.org/10.1007/s12223-009-0013-x
Buckingham J (1993) Dictionary of natural products. CRC Press, London
Running W (1993) Computer software reviews. Chapman and hall dictionary of natural products on CD-ROM. J Chem Inf Model 33:934–935. https://doi.org/10.1021/ci00016a603
Kieser B, Buttner M, Charter K, Hopwood B (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Guo Y, Zheng W, Rong X, Huang Y (2008) A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol 58:149–159. https://doi.org/10.1099/ijs.0.65224-0
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. https://doi.org/10.1128/AEM.00062-07
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. https://doi.org/10.1093/bioinformatics/btp698
Hyatt D, Chen G-L, Locascio PF et al (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. https://doi.org/10.1186/1471-2105-11-119
Blin K, Shaw S, Steinke K et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:81–87. https://doi.org/10.1093/nar/gkz310
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Vinogradov AA, Suga H (2020) Introduction to thiopeptides: biological activity, biosynthesis, and strategies for functional reprogramming. Cell Chem Biol 27(8):1032–1051. https://doi.org/10.1016/j.chembiol.2020.07.003
Chan D, Burrows LL (2021) Thiopeptides: antibiotics with unique chemical structures and diverse biological activities. J Antibiot 74(3):161–175. https://doi.org/10.1038/s41429-020-00387-x
Hrubsky YP, Aravitska OV, Myronovskyy ML et al (2009) N-methyl-N´-nitro-N-nitrosoguanidine and N-methyl-N-nitrosomethylurea influence on antibiotic activity of Streptomyces sioyaensis LV81. Microbiol Biotechnol 2(6):28–35
De Simeis D, Serra S (2021) Actinomycetes: a never-ending source of bioactive compounds-an overview on antibiotics production. Antibiotics (Basel) 10(5):483. https://doi.org/10.3390/antibiotics10050483
Lau RCM, Rinehart KL (1994) Berninamycins B, C, and D, minor metabolites from Streptomyces bernensis. J Antibiot 47(12):1466–1472. https://doi.org/10.7164/antibiotics.47.1466
Chiu ML, Folcher M, Katoh T et al (1999) Broad spectrum thiopeptide recognition specificity of the Streptomyces lividans TipAL protein and its role in regulating gene expression. J Biol Chem 274(29):20578–20586. https://doi.org/10.1074/jbc.274.29.20578
Rong X, Huang Y (2012) Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA–DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol 35(1):7–18. https://doi.org/10.1016/j.syapm.2011.10.004
Matter AM, Hoot SB, Anderson PD et al (2009) Valinomycin biosynthetic gene cluster in Streptomyces: conservation. Ecol Evol PLoS ONE 4(9):e7194. https://doi.org/10.1371/journal.pone.0007194
Malcolmson SJ, Young TS, Ruby JG et al (2013) The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proc Natl Acad Sci US 110(21):8483–8488. https://doi.org/10.1073/pnas.1307111110
Acknowledgements
This work was supported by Grant N/309-2003 from the Ministry of Education and Science of Ukraine to VF, and personal Grant 91657043 from the German Academic Exchange Service (DAAD) to ST. In addition, we would like to thank the scientific reviewers for their detailed and careful reviewing of the manuscript.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
OG conceived and planned the experiments. ST and OG performed the screening of the thiopeptides. MM performed the metabolite extraction and identified the berninamycins. ST performed the phylogenetic and multilocus sequence analyses, the genome sequencing, and the identification of the berninamycin gene cluster. ST and OG both processed the experimental data, performed the analysis, drafted the manuscript, and designed the figures. VF and AL aided in interpreting the results and also worked on the manuscript. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tistechok, S., Myronovskyi, M., Fedorenko, V. et al. Screening of Thiopeptide-Producing Streptomycetes Isolated From the Rhizosphere Soil of Juniperus excelsa. Curr Microbiol 79, 305 (2022). https://doi.org/10.1007/s00284-022-03004-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03004-2