Abstract
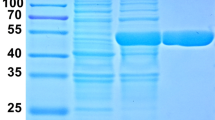
Beta-glucosidase (Bgl) is an enzyme with considerable food, beverage, and biofuel processing potential. However, as many Bgls are inhibited by their reaction end product glucose, their industrial applications are greatly limited. In this study, a novel Bgl gene (Bgl1973) was cloned from Leifsonia sp. ZF2019 and heterologously expressed in E. coli. Sequence analysis and structure modeling revealed that Bgl1973 was 748 aa, giving it a molecular weight of 78 kDa, and it showed high similarity with the glycoside hydrolase 3 (GH3) family Bgls with which its active site residues were conserved. By using pNPGlc (p-nitrophenyl-β-D-glucopyranoside) as substrate, the optimum temperature and pH of Bgl1973 were shown to be 50 °C and 7.0, respectively. Bgl1973 was insensitive to most metal ions (12.5 mM), 1% urea, and even 0.1% Tween-80. This enzyme maintained 60% of its original activity in the presence of 20% NaCl, demonstrating its excellent salt tolerance. Furthermore, it still had 83% residual activity in 1 M of glucose, displaying its outstanding glucose tolerance. The Km, Vmax, and kcat of Bgl1973 were 0.22 mM, 44.44 μmol/min mg, and 57.78 s−1, respectively. Bgl1973 had a high specific activity for pNPGlc (19.10 ± 0.59 U/mg) and salicin (20.43 ± 0.92 U/mg). Furthermore, molecular docking indicated that the glucose binding location and the narrow and deep active channel geometry might contribute to the glucose tolerance of Bgl1973. Our results lay a foundation for the studying of this glucose-tolerant β-glucosidase and its applications in many industrial settings.
Key points
• A novel β-glucosidase from GH3 was obtained from Leifsonia sp. ZF2019.
• Bgl1973 demonstrated excellent glucose tolerance.
• The glucose tolerance of Bgl1973 was explained using molecular docking analysis.






Similar content being viewed by others
Data availability
The authors will make all data underlying the described findings available without restriction upon request.
Code availability
Not applicable.
References
Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272(20):5101–5109. https://doi.org/10.1111/j.1742-4658.2005.04945.x
Arakawa T, Tokunaga M (2004) Electrostatic and hydrophobic interactions play a major role in the stability and refolding of halophilic proteins. Protein Pept Lett 11(2):125–132. https://doi.org/10.2174/0929866043478220
Austin BP, Nallamsetty S, Waugh DS (2009) Hexahistidine-tagged maltose-binding protein as a fusion partner for the production of soluble recombinant proteins in Escherichia coli. Methods Mol Biol 498:157–172. https://doi.org/10.1007/978-1-59745-196-3_11
Bause E, Legler G (1974) Isolation and amino acid sequence of a hexadecapeptide from the active site of beta-glucosidase A3 from Aspergillus wentii. Hoppe Seylers Z Physiol Chem 355(4):438–442. https://doi.org/10.1515/bchm2.1974.355.1.438
Bhatia Y, Mishra S, Bisaria VS (2002) Microbial beta-glucosidases: cloning, properties, and applications. Crit Rev Biotechnol 22(4):375–407. https://doi.org/10.1080/07388550290789568
Bohlin C, Praestgaard E, Baumann MJ, Borch K, Praestgaard J, Monrad RN, Westh P (2013) A comparative study of hydrolysis and transglycosylation activities of fungal β-glucosidases. Appl Microbiol Biotechnol 97(1):159–169. https://doi.org/10.1007/s00253-012-3875-9
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Cao L-c, Wang Z-j, Ren G-h, Kong W, Li L, Xie W, Liu Y-h (2015) Engineering a novel glucose-tolerant β-glucosidase as supplementation to enhance the hydrolysis of sugarcane bagasse at high glucose concentration. Biotechnol Biofuels 8(1):202. https://doi.org/10.1186/s13068-015-0383-z
Caramia S, Gatius AGM, Dal Piaz F, Gaja D, Hochkoeppler A (2017) Dual role of imidazole as activator/inhibitor of sweet almond (Prunus dulcis) beta-glucosidase. Biochem Biophys Rep 10:137–144. https://doi.org/10.1016/j.bbrep.2017.03.007
Chamoli S, Kumar P, Navani NK, Verma AK (2016) Secretory expression, characterization and docking study of glucose-tolerant β-glucosidase from B. subtilis. Int J Biol Macromol 85:425–433. https://doi.org/10.1016/j.ijbiomac.2016.01.001
Chan CS, Sin LL, Chan KG, Shamsir MS, Manan FA, Sani RK, Goh KM (2016) Characterization of a glucose-tolerant β-glucosidase from Anoxybacillus sp DT3-1. Biotechnol Biofuels 9(1):174. https://doi.org/10.1186/s13068-016-0587-x
Chen YA, Zhou Y, Qin YL, Liu DH, Zhao XB (2018) Evaluation of the action of Tween 20 non-ionic surfactant during enzymatic hydrolysis of lignocellulose: pretreatment, hydrolysis conditions and lignin structure. Bioresour Technol 269:329–338. https://doi.org/10.1016/j.biortech.2018.08.119
de Giuseppe PO, Souza TDCB, Souza FHM, Zanphorlin LM, Machado CB, Ward RJ, Jorge JA, Furriel RDM, Murakami MT (2014) Structural basis for glucose tolerance in GH1 beta-glucosidases. Acta Crystallogr D 70:1631–1639. https://doi.org/10.1107/S1399004714006920
Evtushenko LI, Dorofeeva LV, Subbotin SA, Cole JR, Tiedje JM (2000) Leifsonia poae gen nov sp nov isolated from nematode galls on Poa annua and reclassification of “Corynebacterium aquaticum” Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen nov nom rev comb nov and Clavibacter xyli (Davis et al 1984) with two subspecies as Leifsonia xyli Davis et al 1984 gen nov comb nov. Int J Syst Evol Microbiol 50(1):371–380. https://doi.org/10.1099/00207713-50-1-371
Field RA, Haines AH, Chrystal EJT, Luszniak MC (1991) Histidines, histamines and imidazoles as glycosidase inhibitors. Biochem J 274(3):885–889. https://doi.org/10.1042/bj2740885
Florindo RN, Souza VP, Manzine LR, Camilo CM, Marana SR, Polikarpov I, Nascimento AS (2018) Structural and biochemical characterization of a GH3 β-glucosidase from the probiotic bacteria Bifidobacterium adolescentis. Biochimie 148:107–115. https://doi.org/10.1016/j.biochi.2018.03.007
Fusco FA, Fiorentino G, Pedone E, Contursi P, Bartolucci S, Limauro D (2018) Biochemical characterization of a novel thermostable β-glucosidase from Dictyoglomus turgidum. Int J Biol Macromol 113:783–791. https://doi.org/10.1016/j.ijbiomac.2018.03.018
Godse R, Bawane H, Tripathi J, Kulkarni R (2021) Unconventional β-glucosidases: a promising biocatalyst for industrial biotechnology. Appl Biochem Biotechnol 193(9):2993–3016. https://doi.org/10.1007/s12010-021-03568-y
Harnpicharnchai P, Champreda V, Sornlake W, Eurwilaichitr L (2009) A thermotolerant beta-glucosidase isolated from an endophytic fungi Periconia sp with a possible use for biomass conversion to sugars. Protein Expr Purif 67(2):61–69. https://doi.org/10.1016/j.pep.2008.05.022
Huang Y, Busk PK, Grell MN, Zhao H, Lange L (2014) Identification of a β-glucosidase from the Mucor circinelloides genome by peptide pattern recognition. Enzyme Microb Technol 67:47–52. https://doi.org/10.1016/j.enzmictec.2014.09.002
Jabbour D, Klippel B, Antranikian G (2012) A novel thermostable and glucose-tolerant beta-glucosidase from Fervidobacterium islandicum. Appl Microbiol Biotechnol 93(5):1947–1956. https://doi.org/10.1007/s00253-011-3406-0
Kar B, Verma P, den Haan R, Sharma AK (2017) Characterization of a recombinant thermostable β-glucosidase from Putranjiva roxburghii expressed in Saccharomyces cerevisiae and its use for efficient biomass conversion. Process Biochem 63:66–75. https://doi.org/10.1016/j.procbio.2017.08.005
Kaushal G, Rai AK, Singh SP (2021) A novel β-glucosidase from a hot-spring metagenome shows elevated thermal stability and tolerance to glucose and ethanol. Enzyme Microb Technol 145:109764. https://doi.org/10.1016/j.enzmictec.2021.109764
Ketudat Cairns JR, Esen A (2010) beta-Glucosidases. Cell Mol Life Sci 67(20):3389–33405. https://doi.org/10.1007/s00018-010-0399-2
Kim BN, Yeom SJ, Kim YS, Oh DK (2012) Characterization of a β-glucosidase from Sulfolobus solfataricus for isoflavone glycosides. Biotechnol Lett 34(1):125–129. https://doi.org/10.1007/s10529-011-0739-9
Kim IJ, Bornscheuer UT, Nam KH (2022) Biochemical and structural analysis of a glucose-tolerant beta-glucosidase from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum. Molecules 27(1):290. https://doi.org/10.3390/molecules27010290
Kumar S, Stecher G, Tamura K (2016) MEGA7 molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Langston J, Sheehy N, Xu F (2006) Substrate specificity of Aspergillus oryzae family 3 beta-glucosidase. Biochim Biophys Acta 1764(5):972–978. https://doi.org/10.1016/j.bbapap.2006.03.009
Li G, Jiang Y, Fan XJ, Liu YH (2012) Molecular cloning and characterization of a novel β-glucosidase with high hydrolyzing ability for soybean isoflavone glycosides and glucose-tolerance from soil metagenomic library. Bioresour Technol 123:15–22. https://doi.org/10.1016/j.biortech.2012.07.083
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B 2014 The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42(Database issue):D490-D495.https://doi.org/10.1093/nar/gkt1178
Ma Y, Liu X, Yin Y, Zou C, Wang W, Zou S, Hong J, Zhang M (2015) Expression optimization and biochemical properties of two glycosyl hydrolase family 3 beta-glucosidases. J Biotechnol 206:79–88. https://doi.org/10.1016/j.jbiotec.2015.04.016
Méndez-Líter JA, Gil-Muñoz J, Nieto-Domínguez M, Barriuso J, de Eugenio LI, Martínez MJ (2017) A novel, highly efficient β-glucosidase with a cellulose-binding domain: characterization and properties of native and recombinant proteins. Biotechnol Biofuels 10:256. https://doi.org/10.1186/s13068-017-0946-2
Méndez-Líter JA, Nieto-Domínguez M, Fernández de Toro B, González Santana A, Prieto A, Asensio JL, Cañada FJ, de Eugenio LI, Martínez MJ (2020) A glucotolerant β-glucosidase from the fungus Talaromyces amestolkiae and its conversion into a glycosynthase for glycosylation of phenolic compounds. Microb Cell Fact 19(1):127. https://doi.org/10.1186/s12934-020-01386-1
Meng X, Shao Z, Hong Y, Lin L, Li C, Liu Z (2009) A novel pH-stable bifunctional xylanase isolated from a deep-sea microorganism Demequina sp JK4. J Microbiol Biotechnol 19(10):1077–1084
Monteiro LMO, Pereira MG, Vici AC, Heinen PR, Buckeridge MS, Polizeli M (2019) Efficient hydrolysis of wine and grape juice anthocyanins by Malbranchea pulchella β-glucosidase immobilized on MANAE-agarose and ConA-Sepharose supports. Int J Biol Macromol 136:1133–1141. https://doi.org/10.1016/j.ijbiomac.2019.06.106
Monteiro LMO, Vici AC, Pinheiro MP, Heinen PR, de Oliveira AHC, Ward RJ, Prade RA, Buckeridge MS, Polizeli M (2020) A highly glucose tolerant ss-glucosidase from Malbranchea pulchella (MpBg3) enables cellulose saccharification. Sci Rep 10(1):6998. https://doi.org/10.1038/s41598-020-63972-y
Mariano DCB, Leite C, Santos LHS, Marins LF, Machado KS, Werhli AV, Lima LHF, de Melo-Minardi RC (2014) Characterization of glucose-tolerant β-glucosidases used in biofuel production under the bioinformatics perspective: a systematic review. Genet Mol Res 16:(3). https://doi.org/10.4238/gmr16039740
Nielsen H, Tsirigos KD, Brunak S, von Heijne G (2019) A brief history of protein sorting prediction. Protein J 38(3):200–216. https://doi.org/10.1007/s10930-019-09838-3
Park DJ, Lee YS, Choi YL (2013) Characterization of a cold-active β-glucosidase from Paenibacillus xylanilyticus KJ-03 capable of hydrolyzing isoflavones daidzin and genistin. Protein J 32(7):579–584. https://doi.org/10.1007/s10930-013-9520-3
Pyeon HM, Lee YS, Choi YL (2019) Cloning, purification, and characterization of GH3 beta-glucosidase, MtBgl85, from Microbulbifer thermotolerans DAU221. Peer J 7:e7106. https://doi.org/10.7717/peerj.7106
Qu X, Ding B, Li J, Liang M, Du L, Wei Y, Huang R, Pang H (2020) Characterization of a GH3 halophilic β-glucosidase from Pseudoalteromonas and its NaCl-induced activity toward isoflavones. Int J Biol Macromol 164:1392–1398. https://doi.org/10.1016/j.ijbiomac.2020.07.300
Rossi M, Amaretti A, Leonardi A, Raimondi S, Simone M, Quartieri A (2013) Potential impact of probiotic consumption on the bioactivity of dietary phytochemicals. J Agric Food Chem 61(40):9551–9558. https://doi.org/10.1021/jf402722m
Robert X, Gouet P 2014 Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42(Web Server issue):W320-W324.https://doi.org/10.1093/nar/gku316
Salgado JCS, Meleiro LP, Carli S, Ward RJ (2018) Glucose tolerant and glucose stimulated beta-glucosidases - a review. Bioresour Technol 267:704–713. https://doi.org/10.1016/j.biortech.2018.07.137
Sathe SS, Soni S, Ranvir VP, Choudhari VG, Odaneth AA, Lali AM, Chandrayan SK (2017) Heterologous expression and biochemical studies of a thermostable glucose tolerant β-glucosidase from Methylococcus capsulatus (bath strain). Int J Biol Macromol 102:805–812. https://doi.org/10.1016/j.ijbiomac.2017.04.078
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Singh G, Verma AK, Kumar V (2016) Catalytic properties functional attributes and industrial applications of β-glucosidases. 3 Biotech 6(1):3. https://doi.org/10.1007/s13205-015-0328-z
Singhania RR, Patel AK, Pandey A, Ganansounou E (2017) Genetic modification: a tool for enhancing beta-glucosidase production for biofuel application. Bioresour Technol 245:1352–1361. https://doi.org/10.1016/j.biortech.2017.05.126
Sinha SK, Datta S (2016) beta-Glucosidase from the hyperthermophilic archaeon Thermococcus sp is a salt-tolerant enzyme that is stabilized by its reaction product glucose. Appl Microbiol Biotechnol 100(19):8399–8409. https://doi.org/10.1007/s00253-016-7601-x
Su H, Xiao Z, Yu K, Zhang Q, Lu C, Wang G, Wang Y, Liang J, Huang W, Huang X, Wei F (2021) High diversity of β-glucosidase-producing bacteria and their genes associated with Scleractinian corals. Int J Mol Sci 22(7):3523. https://doi.org/10.3390/ijms22073523
Sun J, Wang W, Yao C, Dai F, Zhu X, Liu J, Hao J (2018) Overexpression and characterization of a novel cold-adapted and salt-tolerant GH1 β-glucosidase from the marine bacterium Alteromonas sp L82. J Microbiol 56(9):656–664. https://doi.org/10.1007/s12275-018-8018-2
Sun J, Wang W, Ying Y, Hao J (2020) A novel glucose-tolerant GH1 beta-glucosidase and improvement of its glucose tolerance using site-directed mutation. Appl Biochem Biotechnol 192(3):999–1015. https://doi.org/10.1007/s12010-020-03373-z
Saleh Zada N, Belduz AO, Güler HI, Khan A, Sahinkaya M, Kaçıran A, Ay H, Badshah M, Shah AA, Khan S 2021 Cloning, expression, biochemical characterization, and molecular docking studies of a novel glucose tolerant β-glucosidase from Saccharomonospora sp. NB11. Enzyme Microb Technol 148:109799 https://doi.org/10.1016/j.enzmictec.2021.109799
Teugjas H, Väljamäe P (2013) Selecting β-glucosidases to support cellulases in cellulose saccharification. Biotechnol Biofuels 6(1):105. https://doi.org/10.1186/1754-6834-6-105
Tiwari R, Kumar K, Singh S, Nain L, Shukla P (2016) Molecular detection and environment-specific diversity of glycosyl hydrolase family 1 β-glucosidase in different habitats. Front Microbiol 7:1597. https://doi.org/10.3389/fmicb.2016.01597
Varghese JN, Hrmova M, Fincher GB (1999) Three-dimensional structure of a barley beta-D-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 7(2):179–190. https://doi.org/10.1016/s0969-2126(99)80024-0
Volkov PV, Rozhkova AM, Zorov IN, Sinitsyn AP (2020) Cloning, purification and study of recombinant GH3 family β-glucosidase from Penicillium verruculosum. Biochimie 168:231–240. https://doi.org/10.1016/j.biochi.2019.11.009
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, deBeer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Wu Y, Chi S, Yun C, Shen Y, Tokuda G, Ni J (2012) Molecular cloning and characterization of an endogenous digestive β-glucosidase from the midgut of the fungus-growing termite Macrotermes barneyi. Insect Mol Biol 21(6):604–614. https://doi.org/10.1111/j.1365-2583.2012.01164.x
Wu J, Geng A, Xie R, Wang H, Sun J (2018) Characterization of cold adapted and ethanol tolerant β-glucosidase from Bacillus cellulosilyticus and its application for directed hydrolysis of cellobiose to ethanol. Int J Biol Macromol 109:872–879. https://doi.org/10.1016/j.ijbiomac.2017.11.072
Xia W, Xu X, Qian L, Shi P, Bai Y, Luo H, Ma R, Yao B (2016) Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnol Biofuels 9:147. https://doi.org/10.1186/s13068-016-0560-8
Yang M, Zhang A, Liu B, Li W, Xing J (2011) Improvement of cellulose conversion caused by the protection of Tween-80 on the adsorbed cellulase. Biochem Eng J 56(3):125–129. https://doi.org/10.1016/j.bej.2011.04.009
Yang Y, Zhang X, Yin Q, Fang W, Fang Z, Wang X, Zhang X, Xiao Y (2015) A mechanism of glucose tolerance and stimulation of GH1 β-glucosidases. Sci Rep 5:17296. https://doi.org/10.1038/srep17296
Yin YR, Sang P, Xian WD, Li X, Jiao JY, Liu L, Hozzein WN, Xiao M, Li WJ (2018) Expression and characteristics of two glucose-tolerant GH1 β-glucosidases from Actinomadura amylolytica YIM 77502(T) for promoting cellulose degradation. Front Microbiol 9:3149. https://doi.org/10.3389/fmicb.2018.03149
Yoshida E, Hidaka M, Fushinobu S, Koyanagi T, Minami H, Tamaki H, Kitaoka M, Katayama T, Kumagai H (2010) Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 β-glucosidase from Kluyveromyces marxianus. Biochem J 431(1):39–49. https://doi.org/10.1042/bj20100351
Zada NS, Belduz AO, Güler HI, Sahinkaya M, Khan SI, Saba M, Bektas KI, Kara Y, Kolaylı S, Badshah M, Shah AA, Khan S (2021) Cloning, biochemical characterization and molecular docking of novel thermostable β-glucosidase BglA9 from Anoxybacillus ayderensis A9 and its application in de-glycosylation of Polydatin. Int J Biol Macromol 193:1898–1909. https://doi.org/10.1016/j.ijbiomac.2021.11.021
Zhang C, Zhang L, Wang D, Ma H, Liu B, Shi Z, Ma X, Chen Y, Chen Q (2018) Evolutionary history of the glycoside hydrolase 3 (GH3) family based on the sequenced genomes of 48 plants and identification of jasmonic acid-related GH3 proteins in Solanum tuberosum. Int J Mol Sci 19(7):1850. https://doi.org/10.3390/ijms19071850
Zhang J, Zhao N, Xu J, Qi Y, Wei X, Fan M (2021a) Homology analysis of 35 β-glucosidases in Oenococcus oeni and biochemical characterization of a novel β-glucosidase BGL0224. Food Chem 334:127593. https://doi.org/10.1016/j.foodchem.2020.127593
Zhang P, Zhang R, Sirisena S, Gan R, Fang Z (2021b) Beta-glucosidase activity of wine yeasts and its impacts on wine volatiles and phenolics: a mini-review. Food Microbiol 100:103859. https://doi.org/10.1016/j.fm.2021.103859
Zheng Y, Pan Z, Zhang R, Wang D, Jenkins B (2008) Non-ionic surfactants and non-catalytic protein treatment on enzymatic hydrolysis of pretreated Creeping Wild Ryegrass. Appl Biochem Biotechnol 146(1–3):231–248. https://doi.org/10.1007/s12010-007-8035-9
Zhou J, Zhang R, Shi P, Huang H, Meng K, Yuan T, Yang P, Yao B (2011) A novel low-temperature-active β-glucosidase from symbiotic Serratia sp TN49 reveals four essential positions for substrate accommodation. Appl Microbiol Biotechnol 92(2):305. https://doi.org/10.1007/s00253-011-3323-2
Zmudka MW, Thoden JB, Holden HM (2013) The structure of DesR from Streptomyces venezuelae, a β-glucosidase involved in macrolide activation. Protein Sci 22(7):883–892. https://doi.org/10.1002/pro.2204
Acknowledgements
We thank Dr. David Waugh (National Cancer Institute) for the generous gift of the protein expression vector pET28-HMT and the TEV protease expression strain. In addition, we also thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31970042) and Zhejiang Provincial Natural Science Foundation of China (LY17C200019).
Author information
Authors and Affiliations
Contributions
Y.H. and G.Z.X. contributed to the conception and design of this study and the interpretation of data; Y.H., C.X.W., and R.H.J. performed experiments; Y.H., Q.X.N, Y.W., Q.X.G, and Y.Z.Z. analyzed the data; Y.H. and G.Z.X. wrote the manuscript. All authors contributed to critical revision of the manuscript and gave final approval of the manuscript for publication.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
He, Y., Wang, C., Jiao, R. et al. Biochemical characterization of a novel glucose-tolerant GH3 β-glucosidase (Bgl1973) from Leifsonia sp. ZF2019. Appl Microbiol Biotechnol 106, 5063–5079 (2022). https://doi.org/10.1007/s00253-022-12064-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12064-0