Abstract
Circulating exosomal microRNAs (miRNAs) are valuable biomarker candidates; however, information on the characterization and mutual agreement of commercial kits for circulating exosomal miRNA profiling is scarce. Here, we analyzed the advantages and weaknesses of four commonly used commercial kits for exosomal miRNA profiling and their application to the sample of serum and/or plasma, respectively. NanoSight and Western blotting were conducted to evaluate the efficiency and purity of the isolated exosomes. In our conditions, the size distribution of the isolated particles was appropriate (40–150 nm), and ExoQuick™ Exosome Precipitation Solution (EXQ) generated a relatively high yield of exosomes. Nevertheless, albumin impurity was ubiquitous for all the four kits, and Total Exosome Isolation for serum or plasma (TEI) yielded a relatively pure isolation. We further performed Illumina sequencing combined with RT-qPCR to determine the ability of these kits for miRNA profiling. There was significant correlation of the exosomal miRNA profile and specific miRNAs between kits, but with differences depending on methods. exoRNeasy Serum/Plasma Midi Kit (EXR) and EXQ performed better in the specific exosomal miRNAs recovery. Intraassay CVs for specific miRNA measurement were 0.88–3.82, 1.19–3.77, 0–2.70, and 1.23–9.11% for EXR, TEI, EXQ, and RIBO™ Exosome Isolation Reagent (REI), respectively. In each kit, serum yielded a higher abundance of exosomes and exosomal miRNAs than plasma, yet with more albumin impurity. In conclusion, our data provide some valuable guidance for the methodology of disease biomarker identification of circulation exosomal miRNAs.

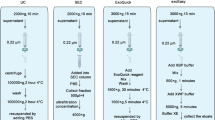
Circulating exosomal microRNAs (miRNAs) are valuable biomarker candidates; however, information on the characterization and mutual agreement of commercial kits for circulating exosomal miRNA profiling is scarce. In this study, we compared four commonly used commercially available kits for exosomal miRNAsextraction and analyzed the advantages and weaknesses of each kit and their application to the sample ofserum and/or plasma





Similar content being viewed by others
References
Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412–20.
Simons M, Raposo G. Exosomes—vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81.
Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93.
Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23:91–7.
Stoorvogel W. Functional transfer of microRNA by exosomes. Blood. 2012;119:646–8.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9.
Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–33.
Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9:358–67.
Pfeffer SR, Grossmann KF, Cassidy PB, Yang CH, Fan M, Kopelovich L, et al. Detection of exosomal miRNAs in the plasma of melanoma patients. J Clin Med. 2015;4:2012–27.
Hosseini M, Khatamianfar S, Hassanian SM, Nedaeinia R, Shafiee M, Maftouh M, et al. Exosome-encapsulated microRNAs as potential circulating biomarkers in colon cancer. Curr Pharm Des. 2017;23:1705–9.
Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–25.
Dolz S, Gorriz D, Tembl JI, Sanchez D, Fortea G, Parkhutik V, et al. Circulating microRNAs as novel biomarkers of stenosis progression in asymptomatic carotid stenosis. Stroke. 2017;48:10–6.
Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2
Boing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. 2014;3
Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394:1253–62.
Nakai W, Yoshida T, Diez D, Miyatake Y, Nishibu T, Imawaka N, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016;6:33935.
Royo F, Diwan I, Tackett MR, Zuniga P, Sanchez-Mosquera P, Loizaga-Iriarte A, Ugalde-Olano A, Lacasa I, Perez A, Unda M, Carracedo A, Falcon-Perez JM. Comparative miRNA analysis of urine extracellular vesicles isolated through five different methods. Cancers (Basel) 2016;8.
Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47:135–8.
Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209.
Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Korvala J, Salo T, Sormunen R, et al. Human saliva-derived exosomes: comparing methods of isolation. J Histochem Cytochem : Off J Histochem Soc. 2015;63:181–9.
Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL. Isolation of biologically-active exosomes from human plasma. J Immunol Methods. 2014;411:55–65.
Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031.
Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–32.
Rood IM, Deegens JK, Merchant ML, Tamboer WP, Wilkey DW, Wetzels JF, et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810–6.
Andreu Z, Rivas E, Sanguino-Pascual A, Lamana A, Marazuela M, Gonzalez-Alvaro I, et al. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J Extracell Vesicles. 2016;5:31655.
Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One. 2017;12:e0170628.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006.
Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–9.
Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7:780–8.
Bobrie A, Colombo M, Krumeich S, Raposo G, Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1
Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61:56–63.
Royo F, Zuniga-Garcia P, Sanchez-Mosquera P, Egia A, Perez A, Loizaga A, et al. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J Extracell Vesicles. 2016;5:29497.
Baranyai T, Herczeg K, Onodi Z, Voszka I, Modos K, Marton N, et al. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One. 2015;10:e0145686.
Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–93.
Chiam K, Wang T, Watson DI, Mayne GC, Irvine TS, Bright T, et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J Gastrointest Surg. 2015;19:1208–15.
Casadei L, Calore F, Creighton CJ, Guescini M, Batte K, Iwenofu OH, et al. Exosome-derived miR-25-3p and miR-92a-3p stimulate liposarcoma progression. Cancer Res. 2017;77:3846–56.
Ebrahimkhani S, Vafaee F, Young PE, Hur SSJ, Hawke S, Devenney E, et al. Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci Rep. 2017;7:14293.
Honegger A, Schilling D, Bastian S, Sponagel J, Kuryshev V, Sultmann H, et al. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015;11:e1004712.
George JN, Thoi LL, McManus LM, Reimann TA. Isolation of human platelet membrane microparticles from plasma and serum. Blood. 1982;60:834–40.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (no. 81472021 and no. 81672102) and Fund of State Key Laboratory of Analytical Chemistry for Life Science (no. 5431ZZXM1601) to C. Zhang, the National Basic Research Program of China (no. 2014CB542300) to C.-Y. Zhang, the National Natural Science Foundation of China (no. 81772282 and no. 81401257) and Foundation of Jiangsu Provincial Medical Youth Talent (QNRC2016893) to C. Wang, and the Scientific Research Foundation of Graduate School of Nanjing University (no. 2016CL09) to M. Ding.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The Ethics Committee of Jinling Hospital (Nanjing, China) approved the present study. Written informed consent was obtained from all participants.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 2460 kb)
Rights and permissions
About this article
Cite this article
Ding, M., Wang, C., Lu, X. et al. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal Bioanal Chem 410, 3805–3814 (2018). https://doi.org/10.1007/s00216-018-1052-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1052-4