Abstract
Background
The poor prognosis and rising incidence of esophageal adenocarcinoma highlight the need for improved detection methods. The potential for circulating microRNAs (miRNAs) as biomarkers in other cancers has been shown, but circulating miRNAs have not been well characterized in esophageal adenocarcinoma. We investigated whether circulating exosomal miRNAs have potential to discriminate individuals with esophageal adenocarcinoma from healthy controls and non-dysplastic Barrett’s esophagus.
Methods
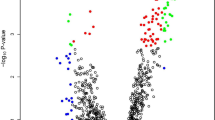
Seven hundred fifty-eight miRNAs were profiled in serum circulating exosomes from a cohort of 19 healthy controls, 10 individuals with Barrett’s esophagus, and 18 individuals with locally advanced esophageal adenocarcinoma. MiRNA expression was assessed using all possible permutations of miRNA ratios per individual. Four hundred eight miRNA ratios were differentially expressed in individuals with cancer compared to controls and Barrett’s esophagus (Mann-Whitney U test, P < 0.05). The 179/408 ratios discriminated esophageal adenocarcinoma from healthy controls and Barrett’s esophagus (linear regression, P < 0.05; area under receiver operating characteristic (ROC) > 0.7, P < 0.05). A multi-biomarker panel (RNU6-1/miR-16-5p, miR-25-3p/miR-320a, let-7e-5p/miR-15b-5p, miR-30a-5p/miR-324-5p, miR-17-5p/miR-194-5p) demonstrated enhanced specificity and sensitivity (area under ROC = 0.99, 95 % CI 0.96–1.0) over single miRNA ratios to distinguish esophageal adenocarcinoma from controls and Barrett’s esophagus.
Conclusions
This study highlights the potential for serum exosomal miRNAs as biomarkers for the detection of esophageal adenocarcinoma.



Similar content being viewed by others
References
Stavrou, E.P., et al., Adenocarcinoma of the oesophagus: incidence and survival rates in New South Wales, 1972–2005. Med J Aust, 2009. 191(6): p. 310–4.
Dubecz, A., et al., Does the Incidence of Adenocarcinoma of the Esophagus and Gastric Cardia Continue to Rise in the Twenty-First Century?-a SEER Database Analysis. J Gastrointest Surg, 2013.
de Jonge, P.J., et al., Barrett’s oesophagus: epidemiology, cancer risk and implications for management. Gut, 2014. 63(1): p. 191–202.
Gordon, L.G., et al., Cost-effectiveness of endoscopic surveillance of non-dysplastic Barrett’s esophagus. Gastrointest Endosc, 2014. 79(2): p. 242–56 e6.
Miranda, K.C., et al., A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell, 2006. 126(6): p. 1203–17.
Sayed, D. and M. Abdellatif, MicroRNAs in development and disease. Physiol Rev, 2011. 91(3): p. 827–87.
Mitchell, P.S., et al., Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A, 2008. 105(30): p. 10513–8.
Wang, J., et al., Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules, 2014. 19(2): p. 1912–38.
Brase, J.C., et al., Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer, 2010. 9: p. 306.
Cheng, L., et al., Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles, 2014. 3.
Koberle, V., et al., Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One, 2013. 8(9): p. e75184.
Ogata-Kawata, H., et al., Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. PLoS One, 2014. 9(4): p. e92921.
Tanaka, Y., et al., Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer, 2013. 119(6): p. 1159–67.
Liao, J., et al., Expression profiling of exosomal miRNAs derived from human esophageal cancer cells by Solexa high-throughput sequencing. Int J Mol Sci, 2014. 15(9): p. 15530–51.
Warnecke-Eberz, U., et al., Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol, 2015.
Zingg, U., et al., Factors associated with postoperative pulmonary morbidity after esophagectomy for cancer. Ann Surg Oncol, 2011. 18(5): p. 1460–8.
Boeri, M., et al., MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A, 2011. 108(9): p. 3713–8.
Bansal, A., et al., Feasibility of mcroRNAs as biomarkers for Barrett’s Esophagus progression: a pilot cross-sectional, phase 2 biomarker study. Am J Gastroenterol, 2011. 106(6): p. 1055–63.
Wu, X., et al., MicroRNA expression signatures during malignant progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer Prev Res (Phila), 2013. 6(3): p. 196–205.
Wijnhoven, B.P., et al., MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. Br J Surg, 2010. 97(6): p. 853–61.
Li, Z., et al., MicroRNA-194 Inhibits the Epithelial-Mesenchymal Transition in Gastric Cancer Cells by Targeting FoxM1. Dig Dis Sci, 2014. 59(9): p. 2145–52.
Dong, P., et al., MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer, 2011. 10: p. 99.
Rahman, M., et al., miR-15b/16-2 regulates factors that promote p53 phosphorylation and augments the DNA damage response following radiation in the lung. J Biol Chem, 2014. 289(38): p. 26406–16.
Weirauch, U., et al., Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia, 2013. 15(7): p. 783–94.
Mogilyansky, E. and I. Rigoutsos, The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ, 2013. 20(12): p. 1603–14.
Sozzi, G., et al., Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol, 2014. 32(8): p. 768–73.
Moldovan, L., et al., Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med, 2014. 18(3): p. 371–90.
Wang, X., et al., Regulation of let-7 and its target oncogenes (Review). Oncol Lett, 2012. 3(5): p. 955–960.
Meyer, S.U., M.W. Pfaffl, and S.E. Ulbrich, Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotechnol Lett, 2010. 32(12): p. 1777–88.
Quidville, V., et al., Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Cancer Res, 2013. 73(7): p. 2247–58.
Appaiah, H.N., et al., Persistent upregulation of U6:SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res, 2011. 13(5): p. R86.
Manterola, L., et al., A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol, 2014. 16(4): p. 520–7.
Author Contributions
All authors participated in aspects of study design, data acquisition and analysis, and preparation of the manuscript.
Funding
This work was supported by a Project grant (APP1022720) and a Centres of Research Excellence grant (APP1040947), both awarded by the National Health and Medical Research Council of Australia, and a Strategic Research Partnership Grant awarded by the Cancer Council of New South Wales.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Predicted probability that each left-out patient in the leave-one-out cross validation has cancer. Each probability was determined using a regression model of a five miRNA ratio panel derived with the sequential testing method utilized in this study. Out of the 18 actual cancer patients in the cohort, the leave-one-out cross validation correctly identified 13 cancer patients from the 18 individuals (highlighted in gray) with the highest predicted probabilities of being a cancer based on the regression model. (DOC 69 kb)
Supplementary Figure 1
Distribution (a) and normality plots (b) of the standardized residuals of the combined five miRNA ratios biomarker panel. (DOC 89 kb)
Supplementary Figure 2
Distribution of the five miRNA ratios in healthy controls, Barrett’s esophagus and esophageal adenocarcinoma. (DOC 395 kb)
Supplementary Figure 3
LOOCV prediction errors for increasing numbers of ratios in the final regression model. Error rate is the number of cancers that were misclassified, as a percentage. (DOC 59 kb)
Rights and permissions
About this article
Cite this article
Chiam, K., Wang, T., Watson, D.I. et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J Gastrointest Surg 19, 1208–1215 (2015). https://doi.org/10.1007/s11605-015-2829-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-015-2829-9