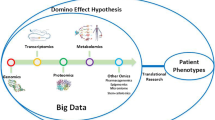
Abstract
Objective
The pathogenesis, diagnosis, and treatment of Sjögren’s syndrome (SS) face many challenges, and there is an urgent need to develop new technologies to improve our understanding of SS.
Methods
By searching the literature published domestically and internationally in the past 20 years, this artical reviewed the research of various omics techniques in SS.
Results
Omics technology provided valuable insights into the pathogenesis, early diagnosis, condition and efficacy evaluation of SS. It is helpful to reveal the pathogenesis of the disease and explore new treatment schemes, which will open a new era for the study of SS.
Conclusion
At present, omics research has made some gratifying achievements, but there are still many uncertainties. Therefore, in the future, we should improve research techniques, standardize the collection of samples, and adopt a combination of multi-omics techniques to jointly study the pathogenesis of SS and provide new schemes for its treatment.

Similar content being viewed by others
Availability of data and materials
Please contact the corresponding author for data requests.
References
Wen Z, Xioamei L, Dong X, Dongzhou L, Jian X, Futao Z, et al. Guidelines for diagnosis and treatment of primary Sjögren’s syndrome. Chin J Intern Med. 2020;59(4):269–76. https://doi.org/10.3760/cma.j.cn112138-20200113-00021. (in Chinese).
Segal BM, Nazmul-Hossain AN, Patel K, Hughes P, Moser KL, Rhodus NL. Genetics and genomics of Sjögren’s syndrome: research provides clues to pathogenesis and novel therapies. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011;111(6):673–80. https://doi.org/10.1016/j.tripleo.2011.01.040. (Epub 2011 Apr 16. PMID: 21497524; PMCID: PMC3095716).
Jianguo W. Sjögren’s syndrome. J Clin Lab. 2022;40(02):138–40. https://doi.org/10.13602/j.cnki.jcls.2022.02.14. (in Chinese).
Dan M, Helin Y, Zewen W, Yazhen S, Liyun Z. Research progress of mesenchymal stem cells and their extracellular vesicles in the treatment of Sjögren’s syndrome. Chin J Rheumatol. 2020;24(09):634–7. https://doi.org/10.3760/cma.j.c141217-20190430-00153. (in Chinese).
Versura P, Giannaccare G, Vukatana G, Mulè R, Malavolta N, Campos EC. Predictive role of tear protein expression in the early diagnosis of Sjögren’s syndrome. Ann Clin Biochem. 2018;55(5):561–70. https://doi.org/10.1177/0004563217750679. (Epub 2018 Jan 30. PMID: 29310465).
Beckman KA, Luchs J, Milner MS. Making the diagnosis of Sjögren’s syndrome in patients with dry eye. Clin Ophthalmol. 2016;10:43–53. https://doi.org/10.2147/opth.s80043. (Epub 2015 Dec 24 PMID: 26766898; PMCID: PMC4699514).
Skarlis C, Marketos N, Mavragani CP. Biologics in Sjögren’s syndrome. Pharmacol Res. 2019;147: 104389. https://doi.org/10.1016/j.phrs.2019.104389. (Epub 2019 Aug 12. PMID: 31415917).
Xiaowen Y, Qin W, Shikai Y, Bin W. Application and thinking of omics technology in the study of Sjögren’s syndrome. Chin J Immunol. 2021;37(15):1895–901. https://doi.org/10.3969/j.issn.1000-484X.2021.15.020.(InChinese).
Gallo A, Baldini C, Teos L, Mosca M, Bombardieri S, Alevizos I. Emerging trends in Sjögren’s syndrome: basic and translational research. Clin Exp Rheumatol. 2012;30(5):779–84 (Epub 2012 Oct 17. PMID: 23009759).
Castro-Santos P, Laborde CM, Diaz-Pena R. Genomics, proteomics and metabolomics: their emerging roles in the discovery and validation of rheumatoid arthritis biomarkers. Clin Exp Rheumatol. 2015;33(2):279–86 (Epub 2015 Jan 8. PMID: 25572119).
Kimoto O, Sawada J, Shimoyama K, Suzuki D, Nakamura S, Hayashi H, et al. Activation of the interferon pathway in peripheral blood of patients with Sjögren’s syndrome. J Rheumatol. 2011;38(2):310–6. https://doi.org/10.3899/jrheum.100486. (Epub 2010 Nov 15. PMID: 21078725).
Lei S, Zhang Y. Identification of the key genes and pathways involved in B cells in primary Sjögren’s syndrome. Bioengineered. 2021;12(1):2055–73. https://doi.org/10.1080/21655979.2021.1930753. (PMID: 34034637; PMCID: PMC8806908).
Lu C, Pi X, Xu W, Qing P, Tang H, Li Y, et al. Clinical significance of T cell receptor repertoire in primary Sjögren’s syndrome. EBioMedicine. 2022;84: 104252. https://doi.org/10.1016/j.ebiom.2022.104252. (Epub 2022 Sep 9. PMID: 36088685; PMCID: PMC9471496).
Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjögren’s syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52(5):1534–44. https://doi.org/10.1002/art.21006. (PMID: 15880807).
Imgenberg-Kreuz J, Rasmussen A, Sivils K, Nordmark G. Genetics and epigenetics in primary Sjögren’s syndrome. Rheumatology (Oxford). 2021;60(5):2085–98. https://doi.org/10.1093/rheumatology/key330. (PMID: 30770922; PMCID: PMC8121440).
Brække-Norheim K, Imgenberg-Kreuz J, Jonsdottir K, Janssen EA, Syvänen AC, Sandling JK, et al. Epigenome-wide DNA methylation patterns associated with fatigue in primary Sjögren’s syndrome. Rheumatology (Oxford). 2016;55(6):1074–82. https://doi.org/10.1093/rheumatology/kew008. (Epub 2016 Mar 10. PMID: 26966136).
Imgenberg-Kreuz J, Almlöf JC, Leonard D, Sjöwall C, Syvänen AC, Rönnblom L, et al. Shared and unique patterns of DNA methylation in systemic lupus erythematosus and primary Sjögren’s syndrome. Front Immunol. 2019;30(10):1686. https://doi.org/10.3389/fimmu. (PMID: 31428085; PMCID: PMC6688520).
Luo X, Peng Y, Chen YY, Wang AQ, Deng CW, Peng LY, et al. Genome-wide DNA methylation patterns in monocytes derived from patients with primary Sjögren’s syndrome. Chin Med J (Engl). 2021;134(11):1310–6. https://doi.org/10.1097/CM9.0000000000001451. (PMID: 33769968; PMCID: PMC8183694).
Miceli-Richard C, Wang-Renault SF, Boudaoud S, Busato F, Lallemand C, Bethune K, et al. Overlap between differentially methylated DNA regions in blood B lymphocytes and genetic at-risk loci in primary Sjögren’s syndrome. Ann Rheum Dis. 2016;75(5):933–40. https://doi.org/10.1136/annrheumdis-2014-206998. (Epub 2015 Jul 16. PMID: 26183421; PMCID: PMC4853580).
Charras A, Konsta OD, Le-Dantec C, Bagacean C, Kapsogeorgou EK, Tzioufas AG, et al. Cell-specific epigenome-wide DNA methylation profile in long-term cultured minor salivary gland epithelial cells from patients with Sjögren’s syndrome. Ann Rheum Dis. 2017;76(3):625–8. https://doi.org/10.1136/annrheumdis-2016-210167. (Epub 2017 Jan 16. PMID: 28093418).
Imgenberg-Kreuz J, Sandling JK, Almlöf JC, Nordlund J, Signér L, Norheim KB, et al. Genome-wide DNA methylation analysis in multiple tissues in primary Sjögren’s syndrome reveals regulatory effects at interferon-induced genes. Ann Rheum Dis. 2016;75(11):2029–36. https://doi.org/10.1136/annrheumdis-2015-208659. (Epub 2016 Feb 8. PMID: 268576; PMCID: PMC5099203).
Spachidou MP, Bourazopoulou E, Maratheftis CI, Kapsogeorgou EK, Moutsopoulos HM, Tzioufas AG, et al. Expression of functional toll-like receptors by salivary gland epithelial cells: increased mRNA expression in cells derived from patients with primary Sjögren’s syndrome. Clin Exp Immunol. 2007;147(3):497–503. https://doi.org/10.1111/j.1365-2249.2006.03311.x. (PMID: 17302899;PMCID: PMC1810489).
Wildenberg ME, Welzen-Coppens JM, Van Helden-Meeuwsen CG, Bootsma H, Vissink A, van Rooijen N, et al. Increased frequency of CD16+ monocytes and the presence of activated dendritic cells in salivary glands in primary Sjögren’s syndrome. Ann Rheum Dis. 2009;68(3):420–6. https://doi.org/10.1136/ard.2008.087874. (Epub 2008 Apr 8. PMID: 18397959).
Witas R, Gupta S, Nguyen CQ. Contributions of major cell populations to Sjögren’s syndrome. J Clin Med. 2020;9(9):3057. https://doi.org/10.3390/jcm9093057. (PMID: 32971904IF; PMCID: PMC7564211).
Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11(11):762–74. https://doi.org/10.1038/nri3070. (PMID: 21984070; PMCID: PMC3947780).
Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steadystate tissues and transport antigen to lymph nodes. Immunity. 2013;39(3):599–610. https://doi.org/10.1016/j.immuni. (Epub 2013 Sep 5. PMID: 24012416; PMCID: PMC3820017).
Wang Y, Xie X, Zhang C, Su M, Gao S, Wang J, et al. Rheumatoid arthritis, systemic lupus erythematosus and primary Sjögren’s syndrome shared megakaryocyte expansion in peripheral blood. Ann Rheum Dis. 2022;81(3):379–85. https://doi.org/10.1136/annrheumdis-2021-220066. (Epub 2021 Aug 30. PMID: 34462261; PMCID: PMC8862024).
Lopes AP, Bekker CPJ, Hillen MR, Blokland SLM, Hinrichs AC, Pandit A, et al. The transcriptomic profile of monocytes from patients with Sjögren’s syndrome is associated with inflammatory parameters and is mimicked by circulating mediators. Front Immunol. 2021;3(12): 701656. https://doi.org/10.3389/fimmu.2021.701656. (PMID: 3441385; PMCID: PMC836872).
Williams AE, Choi K, Chan AL, Lee YJ, Reeves WH, Bubb MR, et al. Sjögren’s syndrome-associated microRNAs in CD14(+) monocytes unveils targeted TGFβ signaling. Arthritis Res Ther. 2016;18(1):95. https://doi.org/10.1186/s13075-016-0987-0. (PMID: 27142093; PMCID: PMC4855899).
Hillen MR, Pandit A, Blokland SLM, Hartgring SAY, Bekker CPJ, van der Heijden EHM, et al. Plasmacytoid DCs from patients with Sjögren’s syndrome are transcriptionally primed for enhanced pro-inflammatory cytokine production. Front Immunol. 2019;4(10):2096. https://doi.org/10.3389/fimmu.2019.02096. (PMID: 31552042; PMCID: PMC6736989).
Lopes AP, Hillen MR, Hinrichs AC, Blokland SL, Bekker CP, Pandit A, et al. Deciphering the role of cDC2s in Sjögren’s syndrome: transcriptomic profile links altered antigen processes with IFN signature and autoimmunity. Ann Rheum Dis. 2023;82(3):374–83. https://doi.org/10.1136/ard-2022-222728. (Epub 2022 Sep 28. PMID: 36171070; PMCID: PMC9933176).
Sun JL, Zhang HZ, Liu SY, Lian CF, Chen ZL, Shao TH, et al. Elevated EPSTI1 promote B cell hyperactivation through NF-κB signalling in patients with primary Sjögren’s syndrome. Ann Rheum Dis. 2020;79(4):518–24. https://doi.org/10.1136/annrheumdis-2019-216428. (Epub 2020 Feb 29. PMID: 32114510).
He C, Yang Y, Chen Z, Liu S, Lyu T, Zeng L, et al. EZH2 promotes T follicular helper cell differentiation through enhancing STAT3 phosphorylation in patients with primary Sjögren’s syndrome. Front Immunol. 2022;20(13): 922871. https://doi.org/10.3389/fimmu.2022.922871. (PMID: 35795677; PMCID: PMC9252457).
Liu S, Yang Y, Zeng L, Wang L, He C, Chen Z, et al. TOX promotes follicular helper T cell differentiation in patients with primary Sjögren’s syndrome. Rheumatology (Oxford). 2023;62(2):946–57. https://doi.org/10.1093/rheumatology/keac304. (PMID: 35713502).
Hinrichs AC, Blokland SLM, Lopes AP, Wichers CGK, Kruize AA, Pandit A, et al. Transcriptome analysis of CCR9+ T helper cells from primary Sjögren’s syndrome patients identifies CCL5 as a novel effector molecule. Front Immunol. 2021;27(12): 702733. https://doi.org/10.3389/fimmu.2021.702733. (PMID: 34386009; PMCID: PMC8354142).
Luo J, Liao X, Zhang L, Xu X, Ying S, Yu M, et al. Transcriptome sequencing reveals potential roles of ICOS in primary Sjögren’s syndrome. Front Cell Dev Biol. 2020;4(8): 592490. https://doi.org/10.3389/fcell.2020.592490. (PMID: 33344450; PMCID: PMC7747463).
Li N, Li L, Wu M, Li Y, Yang J, Wu Y, et al. Integrated bioinformatics and validation reveal potential biomarkers associated with progression of primary Sjögren’s syndrome. Front Immunol. 2021;23(12): 697157. https://doi.org/10.3389/fimmu.2021.697157. (PMID: 3436715; PMCID: PMC8343000).
Oyelakin A, Horeth E, Song EC, Min S, Che M, Marzullo B, et al. Transcriptomic and network analysis of minor salivary glands of patients with primary Sjögren’s syndrome. Front Immunol. 2021;8(11): 606268. https://doi.org/10.3389/fimmu.2020.606268. (PMID: 33488608; PMCID: PMC7821166).
Cross BW, Ruhl S. Glycan recognition at the saliva—oral microbiome interface. Cell Immunol. 2018;333:19–33. https://doi.org/10.1016/j.cellimm.2018.08.008. (Epub 2018 Aug 18. PMID: 30274839; PMCID: PMC6296888).
Li N, Li Y, Hu J, Wu Y, Yang J, Fan H, et al. A link between mitochondrial dysfunction and the immune microenvironment of salivary glands in primary Sjogren’s syndrome. Front Immunol. 2022;14(13): 845209. https://doi.org/10.3389/fimmu.2022.845209. (PMID: 3535993; PMCID: PMC8964148).
Min HK, Moon SJ, Park KS, Kim KJ. Integrated systems analysis of salivary gland transcriptomics reveals key molecular networks in Sjögren’s syndrome. Arthritis Res Ther. 2019;21(1):294. https://doi.org/10.1186/s13075-019-2082-9. (PMID: 31856901; PMCID: PMC6921432).
Verstappen GM, Gao L, Pringle S, Haacke EA, van der Vegt B, Liefers SC, et al. the transcriptome of paired major and minor salivary gland tissue in patients with primary Sjögren’s syndrome. Front Immunol. 2021;6(12): 681941. https://doi.org/10.3389/fimmu.2021.681941. (PMID: 3429533; PMCID: PMC8291032).
Rivière E, Pascaud J, Tchitchek N, Boudaoud S, Paoletti A, Ly B, et al. Salivary gland epithelial cells from patients with Sjögren’s syndrome induce B-lymphocyte survival and activation. Ann Rheum Dis. 2020;79(11):1468–77. https://doi.org/10.1136/annrheumdis-2019-216588. (Epub 2020 Aug 25. PMID: 32843324).
Chen X, Cheng Q, Du Y, Liu L, Wu H. Differential long non-coding RNA expression profile and function analysis in primary Sjogren’s syndrome. BMC Immunol. 2021;22(1):47. https://doi.org/10.1186/s12865-021-00439-3. (PMID: 34284720; PMCID: PMC8293522).
Peng Y, Luo X, Chen Y, Peng L, Deng C, Fei Y, et al. LncRNA and mRNA expression profile of peripheral blood mononuclear cells in primary Sjögren’s syndrome patients. Sci Rep. 2020;10(1):19629. https://doi.org/10.1038/s41598-020-76701-2. (PMID: 33184486; PMCID: PMC7661519).
Shi H, Cao N, Pu Y, Xie L, Zheng L, Yu C. Long non-coding RNA expression profile in minor salivary gland of primary Sjögren’s syndrome. Arthritis Res Ther. 2016;18(1):109. https://doi.org/10.1186/s13075-016-1005-2. (PMID:27188286; PMCID: PMC4869341).
Pertovaara M, Korpela M. ESSPRI and other patient-reported indices in patients with primary Sjogren’s syndrome during 100 consecutive outpatient visits at one rheumatological clinic. Rheumatology (Oxford). 2014;53(5):927–31. https://doi.org/10.1093/rheumatology/ket476. (Epub 2014 Jan 24. PMID: 24464708).
Gottenberg JE, Seror R, Miceli-Richard C, Benessiano J, Devauchelle-Pensec V, Dieude P, et al. Serum levels of beta2-microglobulin and free light chains of immunoglobulins are associated with systemic disease activity in primary Sjogren’s syndrome. Data at enrollment in the prospective ASSESS cohort. PLoS ONE. 2013;8(5):e59868. https://doi.org/10.1371/journal.pone.0059868. (PMID: 23717383; PMCID: PMC3663789).
Alevizos I, Alexander S, Turner RJ, Illei GG. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjögren’s syndrome. Arthritis Rheum. 2011;63(2):535–44. https://doi.org/10.1002/art.30131. (PMID:21280008; PMCID: PMC3653295).
Tandon M, Gallo A, Jang SI, Illei GG, Alevizos I. Deep sequencing of short RNAs reveals novel microRNAs in minor salivary glands of patients with Sjögren’s syndrome. Oral Dis. 2012;18(2):127–31. https://doi.org/10.1111/j.1601-0825.2011.01849.x. (Epub 2011 Sep 6. PMID: 21895886; PMCID: PMC3670135).
Castro I, Carvajal P, Jara D, Aguilera S, Heathcote B, Barrera MJ, et al. Small RNA expression profiling reveals hsa-miR-181d-5p downregulation associated with TNF-α overexpression in Sjögren’s syndrome patients. Front Immunol. 2022;1(13): 870094. https://doi.org/10.3389/fimmu.2022.870094. (PMID: 35432384; PMCID: PMC9010469).
Iwai K, Minamisawa T, Suga K, Yajima Y, Shiba K. Isolation of human salivary extracellular vesicles by iodixanol density gradient ultracentrifugation and their characterizations. J Extracell Vesicles. 2016;17(5):30829. https://doi.org/10.3402/jev.v5.30829. (PMID: 27193612; PMCID: PMC4871899).
Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;14(4):27066. https://doi.org/10.3402/jev.v4.27066. (PMID: 25979354; PMCID: PMC4433489).
Iraci N, Leonardi T, Gessler F, Vega B, Pluchino S. Focus on extracellular vesicles: physiological role and signalling properties of extracellular membrane vesicles. Int J Mol Sci. 2016;17(2):171. https://doi.org/10.3390/ijms17020171. (PMID: 26861302; PMCID: PMC4783905).
Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–83. https://doi.org/10.1083/jcb.201211138. (PMID: 23420871; PMCID: PMC3575529).
Kakan SS, Janga SR, Cooperman B, Craig DW, Edman MC, Okamoto CT, et al. Small RNA deep sequencing identifies a unique miRNA signature released in serum exosomes in a mouse model of Sjögren’s syndrome. Front Immunol. 2020;17(11):1475. https://doi.org/10.3389/fimmu.2020.01475. (PMID: 32849505; PMCID: PMC7396589).
Irudayam JI, Contreras D, Spurka L, Subramanian A, Allen J, Ren S, et al. Characterization of type I interferon pathway during hepatic differentiation of human pluripotent stem cells and hepatitis C virus infection. Stem Cell Res. 2015;15(2):354–64. https://doi.org/10.1016/j.scr.2015.08.003. (Epub 2015 Aug 15. PMID: 26313525; PMCID: PMC4600668).
Tanaka T, Warner BM, Odani T, Ji Y, Mo YQ, Nakamura H, et al. LAMP3 induces apoptosis and autoantigen release in Sjögren’s syndrome patients. 2020;10(1):15169. https://doi.org/10.1038/s41598-020-71669-5(PMID: 32939030; PMCID: PMC7494869).
Nakamura H, Tanaka T, Pranzatelli T, Ji Y, Yin H, Perez P, et al. Lysosome-associated membrane protein 3 misexpression in salivary glands induces a Sjögren’s syndrome-like phenotype in mice. Ann Rheum Dis. 2021;80(8):1031–9. https://doi.org/10.1136/annrheumdis-2020-219649. (Epub 2021 Mar 3. PMID: 33658234; PMCID: PMC8292598).
Cohen P. Signal integration at the level of protein kinases, protein phosphatases and their substrates. Trends Biochem Sci. 1992;17(10):408–13. https://doi.org/10.1016/0968-0004(92)90010-7. (PMID: 1333658).
Sharma K, D’Souza RC, Tyanova S, Schaab C, Wiśniewski JR, Cox J, Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thrbased signaling. Cell Rep. 2014;8(5):1583–94. https://doi.org/10.1016/j.celrep.2014.07.036. (Epub 2014 Aug 21. PMID: 25159151).
Huang S, Zheng F, Liu L, Meng S, Cai W, Zhang C, et al. Integrated proteome and phosphoproteome analyses of peripheral blood mononuclear cells in primary Sjögren syndrome patients. Aging (Albany NY). 2020;13(1):1071–95. https://doi.org/10.18632/aging.202233. (Epub 2020 Dec 3. PMID: 33290261; PMCID: PMC7835054).
Nishikawa A, Suzuki K, Kassai Y, Gotou Y, Takiguchi M, Miyazaki T, et al. Identification of definitive serum biomarkers associated with disease activity in primary Sjögren’s syndrome. Arthritis Res Ther. 2016;18(1):106. https://doi.org/10.1186/s13075-016-1006-1. (PMID: 27180164; PMCID: PMC4868006).
Qiao L, Deng C, Wang Q, Zhang W, Fei Y, Xu Y, et al. Serum clusterin and complement factor H may be biomarkers differentiate primary Sjögren’s syndrome with and without neuromyelitis optica spectrum disorder. Front Immunol. 2019;25(10):2527. https://doi.org/10.3389/fimmu.2019.02527. (PMID: 31708932; PMCID: PMC6823228).
Hjelmervik TO, Jonsson R, Bolstad AI. The minor salivary gland proteome in Sjögren’s syndrome. Oral Dis. 2009;15(5):342–53. https://doi.org/10.1111/j.1601-0825.2009.01531.x. (Epub 2009 Apr 2. PMID: 19364392).
Stea EA, Routsias JG, Samiotaki M, Panayotou G, Papalambros E, Moutsopoulos HM, Tzioufas AG. Analysis of parotid glands of primary Sjögren’s syndrome patients using proteomic technology reveals altered autoantigen composition and novel antigenic targets. Clin Exp Immunol. 2007;147(1):81–9. https://doi.org/10.1111/j.1365-2249.2006.03262.x. (PMID: 17177966; PMCID: PMC1810445).
Giusti L, Baldini C, Bazzichi L, Ciregia F, Tonazzini I, Mascia G, et al. Proteome analysis of whole saliva: a new tool for rheumatic diseases—the example of Sjögren’s syndrome. Proteomics. 2007;7(10):1634–43. https://doi.org/10.1002/pmic.200600783. (PMID: 17436266).
Cecchettini A, Finamore F, Ucciferri N, Donati V, Mattii L, Polizzi E, et al. Phenotyping multiple subsets in Sjögren’s syndrome: a salivary proteomic SWATH-MS approach towards precision medicine. Clin Proteom. 2019;20(16):26. https://doi.org/10.1186/s12014-019-9245-1. (PMID: 31249499; PMCID: PMC6587286).
Sembler-Møller ML, Belstrøm D, Locht H, Pedersen AML. Proteomics of saliva, plasma, and salivary gland tissue in Sjögren’s syndrome and non-Sjögren patients identify novel biomarker candidates. J Proteom. 2020;15(225): 103877. https://doi.org/10.1016/j.jprot.2020.103877. (Epub 2020 Jun 12. PMID: 32540407).
Garreto L, Charneau S, Mandacaru SC, et al. Mapping salivary proteases in Sjögren’s syndrome patients reveals overexpression of dipeptidyl peptidase-4/CD26. Front Immunol. 2021;17(12): 686480. https://doi.org/10.3389/fimmu.2021.686480. (PMID: 34220840; PMCID: PMC8247581).
Aqrawi LA, Chen X, Jensen JL, Morthen MK, Thiede B, Utheim ØA, et al. Severity of clinical dry eye manifestations influences protein expression in tear fluid of patients with primary Sjögren’s syndrome. PLoS ONE. 2018;13(10): e0205762. https://doi.org/10.1371/journal.pone.0205762. (PMID: 30312344; PMCID: PMC6185846).
Aqrawi LA, Galtung HK, Vestad B, Øvstebø R, Thiede B, Rusthen S, et al. Identification of potential saliva and tear biomarkers in primary Sjögren’s syndrome, utilising the extraction of extracellular vesicles and proteomics analysis. Arthritis Res Ther. 2017;19(1):14. https://doi.org/10.1186/s13075-017-1228-x. (PMID: 28122643; PMCID: PMC5264463).
Finamore F, Cecchettini A, Ceccherini E, Signore G, Ferro F, Rocchiccioli S, Baldini C. Characterization of extracellular vesicle cargo in Sjögren’s syndrome through a SWATH-MS proteomics approach. Int J Mol Sci. 2021;22(9):4864. https://doi.org/10.3390/ijms22094864. (PMID: 34064456; PMCID: PMC8124455).
Theander E, Jonsson R, Sjöström B, Brokstad K, Olsson P, Henriksson G. Prediction of Sjögren’s syndrome years before diagnosis and identification of patients with early onset andsevere disease course by autoantibody profiling. Arthritis Rheumatol. 2015;67(9):2427–36. https://doi.org/10.1002/art.39214. (PMID: 26109563).
Di Giorgi N, Cecchettini A, Michelucci E, Signore G, Ceccherini E, Ferro F, et al. Salivary proteomics markers for preclinical Sjögren’s syndrome: a pilot study. Biomolecules. 2022;12(6):738. https://doi.org/10.3390/biom12060738. (PMID: 35740863; PMCID: PMC9221050).
Jovic D, Liang X, Zeng H, Lin L, Xu F, Luo Y. Single-cell RNA sequencing technologies and applications: a brief overview. Clin Transl Med. 2022;12(3): e694. https://doi.org/10.1002/ctm2.694. (PMID: 35352511; PMCID: PMC8964935).
Verstappen GM, Kroese FGM, Bootsma H. T cells in primary Sjogren’s syndrome: targets for early intervention. Rheumatol (Oxford). 2021;60(7):3088–98. https://doi.org/10.1093/rheumatology/kez004. (PMID: 30770920; PMCID: PMC8516500).
Hong X, Meng S, Tang D, Wang T, Ding L, Yu H, et al. Single-cell RNA sequencing reveals the expansion of cytotoxic CD4+ T lymphocytes and a landscape of immune cells in primary Sjögren’s syndrome. Front Immunol. 2021;2(11): 594658. https://doi.org/10.3389/fimmu.2020.594658. (PMID: 33603736; PMCID: PMC7884617).
He Y, Chen R, Zhang M, Wang B, Liao Z, Shi G, et al. Abnormal changes of monocyte subsets in patients with Sjögren’s Syndrome. Front Immunol. 2022;4(13): 864920. https://doi.org/10.3389/fimmu.2022.864920. (PMID: 35309355; PMCID: PMC8931697).
Hou X, Hong X, Ou M, Meng S, Wang T, Liao S, et al. Analysis of gene expression and TCR/B cell receptor profiling of immune cells in primary Sjögren’s syndrome by single-cell sequencing. J Immunol. 2022;209(2):238–49. https://doi.org/10.4049/jimmunol.2100803. (Epub 2022 Jun 15. PMID: 35705251).
Liu J, Gao H, Li C, Zhu F, Wang M, Xu Y, Wu B. Expression and regulatory characteristics of peripheral blood immune cells in primary Sjögren’s syndrome patients using single-cell transcriptomic. iScience. 2022;25(12):105509. https://doi.org/10.1016/j.isci.2022.105509. (PMID: 36425764; PMCID: PMC9678742).
Nguyen CQ, Ogunniyi AO, Karabiyik A, Love JC. Single-cell analysis reveals isotype-specific autoreactive B cell repertoires in Sjögren’s syndrome. PLoS ONE. 2013;8(3): e58127. https://doi.org/10.1371/journal.pone.0058127. (Epub 2013 Mar 13. PMID: 23516437; PMCID: PMC3596347).
Wanchoo A, Voigt A, Sukumaran S, Stewart CM, Bhattacharya I, Nguyen CQ. Single-cell analysis reveals sexually dimorphic repertoires of Interferon-γ and IL-17A producing T cells in salivary glands of Sjögren’s syndrome mice. Sci Rep. 2017;7(1):12512. https://doi.org/10.1038/s41598-017-12627-6. (PMID: 28970488; PMCID: PMC5624952).
Horeth E, Oyelakin A, Song EC, Che M, Bard J, Min S, et al. Transcriptomic and single-cell analysis reveals regulatory networks and cellular heterogeneity in mouse primary Sjögren’s syndrome salivary glands. Front Immunol. 2021;29(12): 729040. https://doi.org/10.3389/fimmu.2021.729040. (PMID: 34912329; PMCID: PMC8666453).
Yan Y, He D. Metabonomics and its application in rheumatology. Chin J Rheumatol. 2023;16(3):203–5. https://doi.org/10.3760/cma.j.issn.1007-7480.2012.03.014. (in Chinese).
Villas-Boas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom Rev. 2005;24(5):613–46. https://doi.org/10.1002/mas.20032. (PMID: 15389842).
Wang K, Li J, Meng D, Zhang Z, Liu S. Machine learning based on metabolomics reveals potential targets and biomarkers for primary Sjogren’s syndrome. Front Mol Biosci. 2022;5(9): 913325. https://doi.org/10.3389/fmolb.2022.913325. (PMID: 36133908; PMCID: PMC9483105).
Kageyama G, Saegusa J, Irino Y, Tanaka S, Tsuda K, Takahashi S, Sendo S, Morinobu A. Metabolomics analysis of saliva from patients with primary Sjögren’s syndrome. Clin Exp Immunol. 2015;182(2):149–53. https://doi.org/10.1111/cei.12683. (Epub 2015 Sep 15. PMID: 26201380; PMCID: PMC4608504).
Li Z, Mu Y, Guo C, You X, Liu X, Li Q, Sun W. Analysis of the saliva metabolic signature in patients with primary Sjögren’s syndrome. PLoS ONE. 2022;17(6): e0269275. https://doi.org/10.1371/journal.pone.0269275. (PMID: 35653354; PMCID: PMC9162338).
Herrala M, Turunen S, Hanhineva K, Lehtonen M, Mikkonen JJW, Seitsalo H, et al. Low-dose doxycycline treatment normalizes levels of some salivary metabolites associated with oral microbiota in patients with primary Sjögren’s syndrome. Metabolites. 2021;11(9):595. https://doi.org/10.3390/metabo11090595. (PMID: 34564411; PMCID: PMC8470364).
Fernández-Ochoa Á, Borrás-Linares I, Quirantes-Piné R, Alarcón-Riquelme ME, Beretta L, Segura-Carretero A, Precisesads Clinical Consortium. Discovering new metabolite alterations in primary sjögren's syndrome in urinary and plasma samples using an HPLC-ESI-QTOF-MS methodology. J Pharm Biomed Anal. 2020;179:112999. https://doi.org/10.1016/j.jpba.2019.112999(Epub 2019 Nov 20. PMID: 31780281).
Yang L, Xiang Z, Zou J, Zhang Y, Ni Y, Yang J. Comprehensive analysis of the relationships between the gut microbiota and fecal metabolome in individuals with primary Sjogren’s syndrome by 16S rRNA sequencing and LC-MS-based metabolomics. Front Immunol. 2022;11(13): 874021. https://doi.org/10.3389/fimmu.2022.874021. (PMID: 35634334; PMCID: PMC9130595).
Zhou Z, Ling G, Ding N, Xun Z, Zhu C, Hua H, et al. Molecular analysis of oral microflora in patients with primary Sjogren’s syndrome by using high-throughput sequencing. PeerJ. 2018;28(6): e5649. https://doi.org/10.7717/peerj.5649. (PMID: 30280027; PMCID: PMC6166617).
Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol. 2018;200(4):525–40. https://doi.org/10.1007/s00203-018-1505-3. (Epub 2018 Mar 23. PMID: 29572583).
Majumdar S, Singh AB. Normal microbial flora of oral cavity. J AdvMed Dent Sci Res. 2014;2(4):62–6.
Zaura E, Brandt BW, Prodan A, Teixeira de Mattos MJ, Imangaliyev S, Kool J, et al. On the ecosystemic network of saliva in healthy young adults. ISME J. 2017;11(5):1218–31. https://doi.org/10.1038/ismej.2016.199. (Epub 2017 Jan 10. PMID: 28072421; PMCID: PMC5475835).
Siddiqui H, Chen T, Aliko A, Mydel PM, Jonsson R, Olsen I. Microbiological and bioinformatics analysis of primary Sjogren’s syndrome patients with normal salivation. J Oral Microbiol. 2016;20(8):31119. https://doi.org/10.3402/jom.v8.31119. (PMID: 27770517; PMCID: PMC5075221).
Zhou S, Cai Y, Wang M, Yang WD, Duan N. Oral microbial flora of patients with Sicca syndrome. Mol Med Rep. 2018;18(6):4895–903. https://doi.org/10.3892/mmr.2018.9520. (Epub 2018 Sep 27. PMID: 30272305; PMCID: PMC6236256).
van der Meulen TA, Harmsen HJM, Bootsma H, Liefers SC, Vich Vila A, Zhernakova A, et al. Dysbiosis of the buccal mucosa microbiome in primary Sjögren’s syndrome patients. Rheumatology (Oxford). 2018;57(12):2225–34. https://doi.org/10.1093/rheumatology/key215. (PMID: 30060225).
Rusthen S, Kristoffersen AK, Young A, Galtung HK, Petrovski BÉ, Palm Ø, et al. Dysbiotic salivary microbiota in dry mouth and primary Sjögren’s syndrome patients. PLoS ONE. 2019;14(6): e0218319. https://doi.org/10.1371/journal.pone.0218319. (PMID: 31211815; PMCID: PMC6581286).
Kim D, Jeong YJ, Lee Y, Choi J, Park YM, Kwon OC, et al. Correlation between salivary microbiome of parotid glands and clinical features in primary sjögren’s syndrome and non-Sjögren’s sicca subjects. Front Immunol. 2022;4(13): 874285. https://doi.org/10.3389/fimmu.2022.874285. (PMID: 35603219; PMCID: PMC9114876).
Sharma D, Sandhya P, Vellarikkal SK, Surin AK, Jayarajan R, Verma A, et al. Saliva microbiome in primary Sjögren’s syndrome reveals distinct set of disease-associated microbes. Oral Dis. 2020;26(2):295–301. https://doi.org/10.1111/odi.13191. (Epub 2020 Jan 10. PMID: 31514257).
Tseng YC, Yang HY, Lin WT, Chang CB, Chien HC, Wang HP, et al. Salivary dysbiosis in Sjögren’s syndrome and a commensal-mediated immunomodulatory effect of salivary gland epithelial cells. NPJ Biofilms Microbiomes. 2021;7(1):21. https://doi.org/10.1038/s41522-021-00192-w. (PMID: 33707430; PMCID: PMC7952914).
Tsigalou C, Stavropoulou E, Bezirtzoglou E. Current insights in microbiome shifts in sjogren’s syndrome and possible therapeutic interventions. Front Immunol. 2018;24(9):1106. https://doi.org/10.3389/fimmu.2018.01106. (PMID: 29881381; PMCID: PMC5976780).
Wang X, Wang J, Guo W, Zhou Y, Sun C, Li Z, Chen L, Pan X. Characteristics of intestinal flora in patients with primary Sjogren’s syndrome. J Southern Med Univ. 2020;40(7):949–57. https://doi.org/10.12122/j.issn.1673-4254.2020.07.06. (in Chinese).
Cano-Ortiz A, Laborda Illanes A, Plaza Andrades I, Membrillo Del Pozo A, Villarrubia Cuadrado A, Rodríguez Calvo de Mora M, et al. Connection between the gut microbiome, systemic inflammation, gut permeability and FOXP3 expression in patients with primary Sjögren’s syndrome. Int J Mol Sci. 2020;21(22):8733. https://doi.org/10.3390/ijms21228733. (PMID: 33228011; PMCID: PMC7699261).
van der Meulen TA, Harmsen HJM, Vila AV, Kurilshikov A, Liefers SC, Zhernakova A, et al. Shared gut, but distinct oral microbiota composition in primary Sjögren’s syndrome and systemic lupus erythematosus. J Autoimmun. 2019;97:77–87. https://doi.org/10.1016/j.jaut.2018.10.009. (Epub 2018 Nov 9. PMID: 30416033).
Wang X, Pang K, Wang J, Zhang B, Liu Z, Lu S, et al. Microbiota dysbiosis in primary Sjögren’s syndrome and the ameliorative effect of hydroxychloroquine. Cell Rep. 2022;40(11): 111352. https://doi.org/10.1016/j.celrep.2022.111352. (PMID: 36103827).
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China Grants (82001741), Science Foundation of Shanxi Health Commission (2023004), and the China Postdoctoral Science Foundation (2023M732147).
Author information
Authors and Affiliations
Contributions
DM and LYZ initiated the project and revised the manuscript. WQH and YYL drafted and wrote the manuscript. RJS, QA, JWZ, and XNG collected the references. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Responsible Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, W., Lu, Y., Shi, R. et al. Application of omics in Sjögren’s syndrome. Inflamm. Res. 72, 2089–2109 (2023). https://doi.org/10.1007/s00011-023-01797-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-023-01797-x