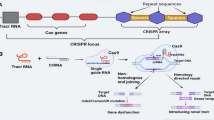
Abstract
Clustered regularly interspaced short palindromic repeat/CRISPR-associated protein (CRISPR-Cas) systems were efficiently used for precise genome editing. CRISPR-Cas systems can generate highly specific double-strand breaks (DSBs) at the target site, and desired sequence modifications can be introduced during the DSB repair process, such as nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) pathways. Among Cas nuclease proteins, Cas9 from Streptococcus pyogenes (SpCas9) is well-studied, and the CRISPR-Cas9 is the most widely used genome editing tool for basic research and crop improvements. It is suitable not only for targeted mutagenesis but also for creating large fragment deletions in plants. In this protocol, we present a step-by-step guide to the CRISPR-Cas9-mediated targeted mutagenesis or large fragment deletions in soybean. Detailed procedures will guide through the essential steps including the design of sgRNAs, construction of CRISPR-Cas9 vectors, Agrobacterium-mediated soybean transformation, and identification of mutant lines.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Gao C (2018) The future of CRISPR technologies in agriculture. Nat Rev 19:275–276
Kim H, Kim ST, Kim SG, Kim JS (2015) Targeted genome editing for crop improvement. Plant Breed Biotechnol 3:283–290
Knott GJ, Doudna JA (2018) CRISPR-Cas guides the future of genetic engineering. Science 361(6405):866–869
Yang N, Wang R, Zhao Y (2017) Revolutionize genetic studies and crop improvement with high-through and genome-scale CRISPR/Cas9 gene editing technology. Mol Plant 10:1141–1143
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Molla KA, Karmakar S, Islam MT (2020) Wide horizons of CRISPR-Cas-derived Technologies for Basic Biology, agriculture, and medicine. In: Islam MT, Bhowmik PK, Molla KA (eds) CRISPR-Cas methods, Springer protocols handbooks. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0616-2_1
Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, Kornepati AVR, Sousa A, Collins MA, Jayaram H, Cullen BR, Bumcrot D (2015) Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol 24(16):257
Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Young JK (2015a) Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 33(12):1293–1298
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, Aryee MJ, Joung JK (2015b) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523(7561):481–485
Xie H, Tang L, He X, Liu X, Zhou C, Liu J, Ge X, Li J, Liu C, Zhao J, Qu J, Song Z, Gu F (2018) SaCas9 requires 50-NNGRRT-30 PAM for sufficient cleavage and possesses higher cleavage activity than SpCas9 or FnCpf1 in human cells. Biotechnol J 14(4):e1700561
Yamano T, Zetsche B, Ishitani R, Zhang F, Nishimasu H, Nureki O (2017) Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Mol Cell 67(4):633–645
Samanta MK, Dey A, Gayen S (2016) CRISPR/Cas9: an advanced tool for editing plant genomes. Transgenic Res 25:561–573
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88
Bao A, Chen H, Chen L, Chen S, Hao Q, Guo W, Qiu D, Shan Z, Yang Z, Yuan S, Zhang C, Zhang X, Liu B, Kong F, Li X, Zhou X, Tran LSP, Cao D (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol 19:131
Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W (2018a) CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol J 16:176–185
Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W (2015) CRISPR/Cas9 mediated genome editing in soybean hairy roots. PLoS One 10:e0136064
Cai Y, Chen L, Sun S, Wu C, Yao W, Jiang B, Han T, Hou W (2018b) CRISPR/Cas9-mediated deletion of large genomic fragments in soybean. Int J Mol Sci 19:3835
Cai Y, Chen L, Zhang Y, Yuan S, Su Q, Sun S, Wu C, Yao W, Han T, Hou W (2020a) Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol J 18:1996–1998
Cai Y, Wang L, Chen L, Wu T, Liu L, Sun S, Wu C, Yao W, Jiang B, Yuan S, Han T, Hou W (2020b) Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol J 18:298–309
Chen L, Cai Y, Qu M, Wang L, Sun H, Jiang B, Wu T, Liu L, Sun S, Wu C, Yao W, Yuan S, Han T, Hou W (2020) Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T. Plant Cell Environ 43:934–944
Cheng Q, Dong L, Su T, Li T, Gan Z, Nan H, Lu S, Fang C, Kong L, Li H, Hou Z, Kou K, Tang Y, Lin X, Zhao X, Chen L, Liu B, Kong F (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol 19:562
Do PT, Nguyen CX, Bui HT, Tran LTN, Stacey G, Gillman JD, Zhang ZJ, Stacey MG (2019) Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol 19:311
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16
Wang L, Sun S, Wu T, Liu L, Sun X, Cai Y, Li J, Jia H, Yuan S, Chen L, Jiang B, Wu C, Hou W, Han T (2020) Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol J 18(9):1869–1881. https://doi.org/10.1111/pbi.13346
Chen L, Cai Y, Liu X, Yao W, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W (2018) Improvement of soybean Agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int J Mol Sci 19:3039
Acknowledgments
This work was supported by the Major Science and Technology Projects of China (2016ZX08010-004), the Ministry of Science and Technology of China (2016YFD0100504), the National Natural Science Foundation of China (31871644), and the CAAS (Chinese Academy of Agriculture Sciences) Agricultural Science and Technology Innovation Project.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Chen, L., Cai, Y., Hou, W. (2021). Generation of Knockout and Fragment Deletion Mutants in Soybean by CRISPR-Cas9. In: Islam, M.T., Molla, K.A. (eds) CRISPR-Cas Methods. Springer Protocols Handbooks. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1657-4_9
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1657-4_9
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1656-7
Online ISBN: 978-1-0716-1657-4
eBook Packages: Springer Protocols