Abstract
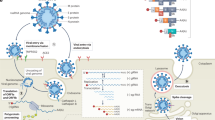
The COVID-19 pandemic is presently the major threat to human society and health due to its high infectivity and mortality rates. To date, this pandemic has resulted in more than 1.5 million deaths globally, affecting more than 200 countries. Phylogenetic analysis of the SARS-CoV-2 genome revealed its striking homology with the bat-derived coronavirus strains, thus confirming the zoonotic origin of the virus. SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE2) receptors expressed on the surface of the host cells, leading to endocytosis of the receptor, followed by the replication of the viral RNA, packaging, assembly, and release of the progeny viruses. This leads to the systemic infection in the host body and the shredding of the virus, causing its transmission to a new host. The extent of infection in the host cells depends on the expression of ACE2 expression and hyperactivation of the immune system to generate a cocktail of inflammatory cytokines, also referred to as the cytokine storm. This inflammatory response can cause severe damage to the lung tissues.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Gorbalenya AE et al (2020) The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5(4):536–544. https://doi.org/10.1038/s41564-020-0695-z
Chan JF-W et al (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) 395(10223):514–523
Lu H, Stratton CW, Tang Y-W (2020) Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol 92(4):401–402
Burki TK (2020) Coronavirus in China. Lancet Respir Med 8(3):238
WHO (2020) WHO Director-General’s opening remarks at the media briefing on COVID-19, 11 Mar 2020. Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19%2D%2D-11-march-2020
Burrell CJ, Howard CR, Murphy FA (2016) Fenner and White’s medical virology, 5th edn. Academic Press, Amsterdam
Woo PCY et al (2012) Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86(7):3995–4008. Available from: http://europepmc.org/abstract/MED/22278237
Wu F et al (2020) A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269
Zhou P et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. https://doi.org/10.1038/s41586-020-2012-7
Wu A et al (2020) Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27(3):325–328
Tanner JA et al (2003) The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J Biol Chem 278(41):39578–39582. Available from: http://europepmc.org/abstract/MED/12917423
Krichel B et al (2020) Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem J 477(5):1009–1019
Van Boheemen S et al (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3(6):e00473-12
Wang Q et al (2020) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181(4):894–904.e9. Available from: http://www.sciencedirect.com/science/article/pii/S009286742030338X
Nieto-Torres JL et al (2011) Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 415(2):69–82
Kirchdoerfer RN, Ward AB (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10(1):2342. https://doi.org/10.1038/s41467-019-10280-3
China Center for Disease Control and Prevention (2020) China CDC detects a large number of new coronaviruses at Huanan Seafood Market in Wuhan. Available from: http://www.chinacdc.cn/yw_9324/202001/t20200127_211469.html. Accessed 20 Feb 2020
Rothe C et al (2020) Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med 382:970–971
Bai Y et al (2020) Presumed asymptomatic carrier transmission of COVID-19. JAMA 323(14):1406–1407
Oran DP, Topol EJ (2020) Prevalence of asymptomatic SARS-CoV-2 infection. Ann Internal Med 173(5):362–367. https://doi.org/10.7326/M20-3012
Li Q et al (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382(13):1199–1207
Netea MG et al (2020) Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 infection. Cell 181(5):969–977. Available from: http://www.sciencedirect.com/science/article/pii/S0092867420305079
Biswas N, Majumder P (2020) Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type. Indian J Med Res 151(5):450–458. Available from: http://www.ijmr.org.in/preprintarticle.asp?id=284484;type=0
Bhattacharyya C et al (2020) Global spread of SARS-CoV-2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. BioRxiv. Available from: http://biorxiv.org/content/early/2020/05/05/2020.05.04.075911.abstract
Hu J et al (2020) The D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity and decreases neutralization sensitivity to individual convalescent sera. BioRxiv. Available from: https://www.biorxiv.org/content/early/2020/06/20/2020.06.20.161323
Korber B et al (2020) Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182(4):812–827.e19. Available from: https://pubmed.ncbi.nlm.nih.gov/32697968
Shang J et al (2020) Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci 117(21):11727–11734. Available from: https://www.pnas.org/content/117/21/11727
Hoffmann M et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271–280.e8. Available from: http://www.sciencedirect.com/science/article/pii/S0092867420302294
Tang T et al (2020) Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir Res 178:104792
Banerjee AK et al (2020) SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. Available from: http://www.sciencedirect.com/science/article/pii/S0092867420313106
Jia HP et al (2009) Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol 297(1):L84–L96
Yang Y et al (2020) SARS-CoV-2: characteristics and current advances in research. Virol J 17(1):117. https://doi.org/10.1186/s12985-020-01369-z
Wu D, Yang XO (2020) TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect (Wei Mian Yu Gan Ran Za Zhi) 53(3):368–370
Cao Y et al (2020) Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov 6(1):11. https://doi.org/10.1038/s41421-020-0147-1
Renieri A et al (2020) ACE2 variants underlie interindividual variability and susceptibility to COVID-19 in Italian population. MedRxiv. Available from: https://www.medrxiv.org/content/early/2020/04/06/2020.04.03.20047977
Stawiski EW et al (2020) Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. BioRxiv. Available from: https://www.biorxiv.org/content/early/2020/04/10/2020.04.07.024752
Hussain M et al (2020) Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J Med Virol 92(9):1580–1586. https://doi.org/10.1002/jmv.25832
Russo R et al (2020) Genetic analysis of the coronavirus SARS-CoV-2 host protease TMPRSS2 in different populations. Front Genet 11:872
Ozono S et al (2020) Naturally mutated spike proteins of SARS-CoV-2 variants show differential levels of cell entry. BioRxiv. Available from: https://www.biorxiv.org/content/early/2020/06/26/2020.06.15.151779
Asselta R et al (2020) ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging 12(11):10087–10098. https://doi.org/10.18632/aging.103415
Torre-Fuentes L et al (2020) ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J Med Virol. https://doi.org/10.1002/jmv.26319
The Severe Covid-19 GWAS Group (2020) Genomewide Association Study of severe Covid-19 with respiratory failure. N Engl J Med 383(16):1522–1534. https://doi.org/10.1056/NEJMoa2020283
Zeberg H, Pääbo S (2020) The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. https://doi.org/10.1038/s41586-020-2818-3
Bastard P et al (2020) Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science:eabd4585. Available from: http://science.sciencemag.org/content/early/2020/09/23/science.abd4585.abstract
Tu Y-F et al (2020) A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci 21(7):2657
Yin W et al (2020) Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368(6498):1499–1504. Available from: http://science.sciencemag.org/content/368/6498/1499.abstract
Beigel JH et al (2020) Remdesivir for the treatment of Covid-19—final report. N Engl J Med. https://doi.org/10.1056/NEJMoa2007764
Cai Q et al (2020) Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering (Beijing, China) 6:1192–1198
Caly L et al (2020) The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res 178:104787. Available from: http://www.sciencedirect.com/science/article/pii/S0166354220302011
Hung IF-N et al (2020) Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 395(10238):1695–1704. https://doi.org/10.1016/S0140-6736(20)31042-4
Zoufaly A et al (2020) Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir Med. https://doi.org/10.1016/S2213-2600(20)30418-5
Wang X et al (2020) The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov 6(1):28. https://doi.org/10.1038/s41421-020-0169-8
Nojomi M et al (2020) Effect of arbidol on COVID-19: a randomized controlled trial. Available from: http://europepmc.org/abstract/PPR/PPR217076
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ghosh, A., Bhattacharyya, C., Biswas, N.K., Das, A. (2021). Underpinning the Rudimentary/Underlying Mechanisms Involved in the Pathogenesis of SARS-CoV-2 (COVID-19) in Human Lung Cells. In: Dua, K., Löbenberg, R., Malheiros Luzo, Â.C., Shukla, S., Satija, S. (eds) Targeting Cellular Signalling Pathways in Lung Diseases. Springer, Singapore. https://doi.org/10.1007/978-981-33-6827-9_25
Download citation
DOI: https://doi.org/10.1007/978-981-33-6827-9_25
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-33-6826-2
Online ISBN: 978-981-33-6827-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)