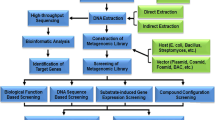
Abstract
Lignocellulose is considered as one of the most copious biopolymer accessible on this planet. Lignocellulosic hydrolysis which yields sugar and phenolics is a must for fermentation processes and pilot scale production of value added products. Cellulases are the class of enzymes which are mainly produced by fungi and bacteria and help in cellulose hydrolysis by acting on the β-1,4 linkages of cellulosic chains. The microbial cellulases have been found to be used in several industries such as biofuel, food, brewing, textile and laundry. Recently, functional metagenomics have been found to be an important strategy for the discovery of cellulose genes. However, the efficiency of such techniques for enzyme discovery from environmental metagenomes is not sufficient to meet the increasing industrial demands. Scientific and industrial advancements, role of metagenomics and future scenario related to the application of several cellulase pertaining to different industries will be discussed in this chapter.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Acharya S, Chaudhary A (2012) Bioprospecting thermophiles for cellulase production: a review. Braz J Microbiol 843:44–856
Adrio JL, Demain AL (2014) Microbial enzymes: tools for biotechnological processes. Biomol Ther 4:117–139
Alvarez TM, Paiva JH, Ruiz DM et al (2013) Structure and function of a novel cellulase 5 from sugarcane soil metagenome. PLoS One 8:e83635
Arja MO (2007) Cellulases in the textile industry. In: Polaina J, MacCabe AP (eds) Industrial enzymes. Springer, Dordrecht, pp 51–63
Bhat MK (2000) Cellulases and related enzymes in biotechnology. Biotechnol Adv 18:355–383
Cheema S, Bassas-Galia M, Sarma PM, Lal B, Arias S (2012) Exploiting metagenomic diversity for novel polyhydroxyalkanoate synthases: production of a terpolymer poly (3-hydroxybutyrate-co-3-hydroxyhexanoate-co-3-hydroxyoctanoate) with a recombinant Pseudomonas putida strain. Bioresour Technol 103:322–328
Çinar I (2005) Effects of cellulase and pectinase concentrations on the colour yield of enzyme extracted plant carotenoids. Process Biochem 40:945–949
De Faveri D, Aliakbarian B, Avogadro M, Perego P, Converti A (2008) Improvement of olive oil phenolics content by means of enzyme formulations: effect of different enzyme activities and levels. Biochem Eng J 41:149–156
De Vries M, Schöler A, Ertl J, Xu Z, Schloter M (2015) Metagenomic analyses reveal no differences in genes involved in cellulose degradation under different tillage treatments. FEMS Microbiol Eco 91:69
Demain AL, Newcomb M, Wu JHD (2005) Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154
Dienes D, Egyházi A, Réczey K (2004) Treatment of recycled fiber with Trichoderma cellulases. Ind Crop Prod 20:11–21
Duan CJ, Xian L, Zhao GC et al (2009) Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol 107:245–256
Freier D, Mothershed CP, Wiegel J (1988) Characterization of Clostridium thermocellum JW20. Appl Environ Microbiol 54:204–211
Gilbert JA, Dupont CL (2011) Microbial metagenomics: beyond the genome. Ann Rev Marine Sci 3:347–371
Gong X, Gruniniger RJ, Forster RJ, Teather RM, McAllister TA (2013) Biochemical analysis of a highly specific, pH stable xylanase gene identified from a bovine rumen-derived metagenomic library. Appl Microbiol Biotechnol 97:2423–2431
Hamilton-Brehm SD, Mosher JJ, Vishnivetskaya T, Podar M et al (2010) Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl Environ Microbiol 76:1014–1020
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5:R245–R249
Hess M, Sczyrba A, Egan R et al (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467
Howell CR (2003) Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis 87:4–10
Jiménez DJ, de Lima Brossi MJ, Schückel J, Kračun SK, Willats WGT, van Elsas JD (2016) Characterization of three plant biomass-degrading microbial consortia by metagenomics- and metasecretomics-based approaches. Appl Microbiol Biotechnol 100:10463–10477
Jorgensen KS (2007) In situ bioremediation. Adv Appl Microbiol 61:285–305
Juturu V, Wu JC (2014) Microbial cellulases: engineering, production and applications. Renew Sust Energ Rev 33:188–120
Kanafusa-Shinkai S, Wakayama J, Tsukamoto K, Hayashi N, Miyazaki Y, Ohmori H, Tajima K, Yokoyama H (2013) Degradation of microcrystalline cellulose and non-pretreated plant biomass by a cell-free extracellular cellulase/hemicellulase system from the extreme thermophilic bacterium. J Biosci Bioeng 115:64–70
Kanokratana P, Eurwilaichitr L, Pootanakit K, Champreda V (2015) Identification of glycosyl hydrolases from a metagenomic library of microflora in sugarcane bagasse collection site and their cooperative action on cellulose degradation. J Biosci Bioeng 119:384–391
Karmakar M, Ray RR (2011) Current trends in research and application of microbial cellulases. Res J Microbiol 6:41–53
Kato S, Haruta S, Cui ZJ, Ishii M, Yokota A, Igarashi Y (2004) Clostridium straminisolvens sp. nov., a moderately thermophilic, aerotolerant and cellulolytic bacterium isolated from a cellulose-degrading bacterial community. Int J Syst Evol Microbiol 54:2043–2047
Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y (2005) Effective cellulose degradation by a mixed-culture system composed of a cellulolytic Clostridium and aerobic non-cellulolytic bacteria. FEMS Microbiol Ecol 51:133–142
Knapp JS (1985) Biodegradation of cellulose and lignins. Comprehen Biotechnol 4:835
Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B (2009) Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels 2:19
Kuhad RC, Gupta R, Singh A (2011) Microbial cellulases and their industrial applications. Enzyme Res 2011:280696
Kung L Jr, Kreck EM, Tung RS, Hession AO, Sheperd AC, Cohen MA, Swain HE Leedle JA (1997) Effects of a live yeast culture and enzymes on in vitro ruminal fermentation and milk production of dairy cows. J Dairy Sci 80:2045–2051
Lee M, Lee C, Oh T, Song JK, Yoon J (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72:7406–7409
López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P (2016) Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep 6:25279
Lynd LR, Weimer PJ, Van Zyl WH, Isak S, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
McDonald JE, Houghton JNI, Rooks D, Allison HE, McCarthy AJ (2012) The microbial ecology of anaerobic cellulose degradation in municipal waste landfill sites: evidence of a role for fibrobacters. Environ Microbiol 14:1077–1087
Milala MA, Shugaba A, Gidado A, ENe AC, Wafar JA (2005) Studies on the use of agricultural wastes for cellulase enzyme production by Aspergillus niger. Res J Agric Biol Sci 1:325–328
Phitsuwan P, Laohakunjit N, Kerdchoechuen O, Kyu KL, Ratanakhanokchai K (2012) Present and potential applications of cellulases in agriculture, biotechnology, and bioenergy. Folia Microbiol 58:163–176
Pottkämper J, Barthen P, Ilmberger N, Schwaneberg U, Schenk A, Schulte M, Streit WR (2009) Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids. Green Chem 11:957
Rai P, Majumdar G, Gupta SD, De S (2007) Effect of various pretreatment methods on permeate flux and quality during ultrafiltration of mosambi juice. J Food Eng 78:561–568
Ransom-Jones E, McCarthy AJ, Haldenby S, Doonan J, McDonald JE (2017) Lignocellulose-degrading microbial communities in landfill sites represent a repository of unexplored biomass-degrading diversity. mSphere 2:e00300–e00317
Rapp P, Beerman (1991) Bacterial cellulases. In: Weimer CH (ed) Biosynthesis and degradation of cellulose. Marcel Dekker, New York, pp 535–595
Resch MG, Donohoe BS, Baker JO, Decker SR, Bayer EA, Beckham GT, Himmel ME (2013) Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction. Energy Environ Sci 6:1858–1867
Schallmey M, Ly A, Wang C et al (2011) Harvesting of novel polyhydroxyalkanaote (PHA) synthase encoding genes from a soil metagenome library using phenotypic screening. FEMS Microbiol Lett 321:150–156
Schwarz WH (2001) The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol 56:634–649
Sharada R, Venkateswarlu G, Venkateswar S, AnandRao M (2014) Applications of cellulases—review. Int J Pharm Chem Biol Sci 4:424–437
Singh A, Kuhad RC, Ward OP (2007) Industrial application of microbial cellulases. In: Kuhad RC, Singh A (eds) Lignocellulose biotechnology: future prospects. I.K. International Publishing House, New Delhi, pp 345–358
Sizova MV, Izquierdo JA, Panikov NS, Lynd LR (2011) Cellulose- and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl Environ Microbiol 77:2282–2291
Thornbury M, Sicheri J, Slaine P, Getz LJ, Finlayson-Trick E, Cook J et al (2019) Characterization of novel lignocellulose-degrading enzymes from the porcupine microbiome using synthetic metagenomics. PLoS One 14:e0209221
Voget S, Steele HL, Streit WR (2006) Characterization of a metagenome-derived halotolerant cellulose. J Biotechnol 126:26–36
Waghunge RR, Rahul MS, Ambalal NS (2016) Trichoderma: a significant fungus for agriculture and environment. Afr J Agr Res 11:1952–1965
Wang C, Dong D, Wang H, Müller K, Qin Y, Wang H, Wu W (2016) Metagenomic analysis of microbial consortia enriched from compost: new insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol Biofuels 9:22
Wang L, Jiang X, Xu H, Zhang Y (2018) Metagenomic analysis of cellulose and volatile fatty acids metabolism by microorganisms in cow rumen. BioRxiv 414961
Wilhelm RC, Singh R, Eltis LD, Mohn WW (2019) Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J 13:413–429
Wong MT, Wang W, Couturier M et al (2017) Comparative metagenomics of cellulose- and poplar hydrolysate-degrading microcosms from gut microflora of the Canadian Beaver (Castor canadensis) and North American Moose (Alces americanus) after long-term enrichment. Front Microbiol 8:2504
World Enzymes (2011) Industry study with forecasts for 2015 & 2020. The Freedonia Group, USA, pp 1–376
Yeh Y, Chang SC, Kuo H, Tong C, Yu S, Ho TD (2013) A metagenomic approach for the identification and cloning of an endoglucanase from rice straw compost. Gene 519:360–366
Zhang L, Chung J, Jiang Q, Sun R, Zhang J, Zhong Y, Ren N (2017) Characteristics of rumen microorganisms involved in anaerobic degradation of cellulose at various pH values. RSC Adv 7:40303–40310
Zverlov VV, Schwarz WH (2008) Bacterial cellulose hydrolysis in anaerobic environmental subsystems- Clostridium thermocellum and Clostridium stercorarium, thermophilic plant fiber degraders. Ann NY Acad Sci 1125:298–307
Acknowledgement
PJ is thankful to Amity University, Kolkata for providing the infrastructure facilities. VK would also like to acknowledge Department of Science and Technology (DST), SERB, GoI for the financial support under Startup Research Scheme (File No.: SRG/2019/001279).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, V., Jha, P. (2020). Role of Metagenomics in Discovery of Industrially Important Cellulase. In: Shrivastava, S. (eds) Industrial Applications of Glycoside Hydrolases . Springer, Singapore. https://doi.org/10.1007/978-981-15-4767-6_10
Download citation
DOI: https://doi.org/10.1007/978-981-15-4767-6_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4766-9
Online ISBN: 978-981-15-4767-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)