Abstract
The cardiac conduction system (CCS) consists of distinctive components that initiate and conduct the electrical impulse required for the coordinated contraction of the cardiac chambers. The development of the CCS involves complex regulatory networks of transcription factors that act in stage, tissue and dose-dependent manners. As disrupted function or expression of these factors may lead to disorders in the development or function of components of the CCS associated with heart failure and sudden death, it is crucial to understand the molecular and cellular mechanisms underlying their complex regulation. Here, we discuss the regulation of genes driving CCS-specific gene expression and demonstrate the complexity of the mechanisms governing their regulatory networks. The three-dimensional conformation of chromatin has recently been recognized as an important regulatory layer, shaping the genome in regulatory domains and physically wiring gene promoters to their regulatory sequences. Knowledge of the mechanisms by which distal-acting regulatory sequences exert their function to drive tissue-specific gene expression and understanding how the three-dimensional chromatin landscape is involved in this regulation will increase our understanding of how disease-associated genomic variation affects the function of such sequences.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Heart development
- Conduction system
- Sinus node
- Atrioventricular node
- Transcriptional regulation
- Functional genomics
- Patterning
- Pacemaker
1 Introduction
The cardiac conduction system (CCS) initiates and propagates the electrical impulse that is required for the rhythmic and synchronized contraction of the heart. The impulse initiates in the sinoatrial node (SAN) and is rapidly propagated through the atria, thereby activating the contraction of the atrial myocardium. The impulse is then propagated through the atrioventricular node (AVN), the only electrical connection between the atria and ventricles. The AVN delays conduction of the impulse, allowing for the atrial contraction and ventricular filling to complete before the ventricles contract. Further propagation of the impulse to the fast-conducting atrioventricular bundle (AVB), bundle branches (BBs) and Purkinje network causes the depolarization of the ventricular myocardium, leading to ventricular contraction. The development of the CCS is regulated by transcription factors that act in strictly stage, tissue and dose-dependent manners [1, 2]. A disruption in the function or expression of these factors could lead to disorders in the development or function of the CCS that can lead to lethal arrhythmias and heart failure. Knowledge of the mechanisms underlying regulation of genes involved in CCS development is therefore crucial.
2 Genetic Pathways Controlling SAN and AVC Development
The heart is the first organ to form during embryonic development and starts as a primitive, linear tube with the inflow region at the caudal side and the outflow region at the cranial side. The slow conductive properties of the embryonic muscle cells within the primitive tube at this stage and dominant pacemaker activity at the caudal end cause a slow peristaltic pattern of contraction along the tube to propagate the blood. At this stage, the entire sinus venosus acts as pacemaker, characterized by the expression of the pacemaker channel Hcn4, a member of the family of channels responsible for the hyperpolarization-activated current if that is crucial for the pacemaker potential [3, 4]. The heart tube elongates by the addition of rapidly proliferating progenitor cells that differentiate to cardiac muscle. This implies that cells added to the inflow tract will acquire dominant pacemaker activity. With further development, specific regions in the heart tube start to divide rapidly and activate a working myocardial gene program, resulting in the ballooning of the primitive atrium and ventricle. Concomitant with the ballooning of the primitive atria is the formation of the sinus venosus including the SAN. Dominant pacemaker activity will gradually be confined to the SAN at the junction of the sinus venosus and atrium.
The transcriptional activator Tbx5 is required for the sinus venosus expression of Shox2 [5], a homeobox transcription factor necessary for SAN formation and function [6, 7]. Shox2 represses cardiac homeobox transcription factor Nkx2-5 [6]. In the chambers, Nkx2-5 activates chamber-specific genes including Nppa and high-conductance gap junction subunit-encoding genes Gja5 (Cx40) and Gja1 (Cx43), whereas it represses SAN/CCS-specific genes Hcn4 and T-box transcription factor Tbx3 [8]. Tbx3 is required for the formation of the SAN by directly repressing atrial myocardial genes Gja5, Gja1 and Nppa to prevent atrialization of the SAN and indirectly activating Hcn4 and other SAN genes [9, 10].
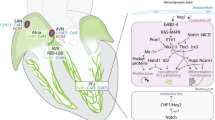
During ballooning of the primitive cardiac chambers, the region in between the atria and ventricles does not proliferate and forms a constriction, the atrioventricular canal (AVC). Bmp2 expression in the AVC activates the expression of Tbx3 and Tbx2 [11]. Together with Msx2, these T-box factors repress the working myocardial gene program in the AVC and AVC-derived AVN [12, 13] and stimulate the pacemaker gene program and the program required for the formation of the AV cushions. Within the AVC, Tbx2 and Tbx3 interact with Nkx2-5 to repress genes that are activated by Nkx2-5 and Tbx5 in the working myocardium of the atria and ventricles [14–16]. Tbx2 and Tbx3 thus suppress working myocardial differentiation of the AVC, thereby causing the retention of the primitive phenotype of slow conduction and low rates of proliferation, providing a primitive morphological and functional constriction in between the atrial and ventricular chambers. Other factors that regulate the formation of the AVC and its border with the chamber myocardium include Wnts, acting upstream of Bmp2; Hey1 and Hey2, Notch target genes expressed in the chambers that suppress Tbx2 [17]; Tbx20, which represses BMP-mediated activation of Tbx2 in the chambers [18]; and Gata4/6, which act in complex with Smads and histone acetyltransferases (HATs) to activate AVC-specific enhancers in the AVC and with histone deacetylases (HDACs) and Hey1/2 to suppress these enhancers in the chambers [19]. The resulting pattern of conduction—fast in the atria, slow in the AVC and fast in the ventricles—results in the alternating contraction pattern of the chambers and an ECG that resembles the adult ECG (Fig. 38.1a).
Schematic overview of cardiac development. (a) The early heart tube has a primitive phenotype of slow conduction, represented by a sinusoidal ECG. With further development, regions at the outer curvatures of the primary heart tube expand and obtain a working myocardium phenotype of fast conduction (grey). The sinus venosus (sv), AVC, outflow tract (oft) and inner curvatures retain their primitive pacemaker-like phenotype and slow conductivity (purple). A more mature ECG can be derived from these hearts. Eventually, these non-chamber myocardial regions will give rise to the mature conduction system components. (b) The transcriptional repressors Tbx2 and Tbx3 compete with the transcriptional activator Tbx5 to regulate their target genes. In the AVC, expression of chamber myocardium genes like Nppa is actively repressed by Tbx2 and Tbx3, whereas in the developing chambers Tbx5 activates these genes. Transcriptional activation in the AVC is regulated by GATA binding site-dependent histone modifications which render the chromatin more (e.g. HATs) or less (e.g. HDACs) accessible for transcription factors to bind their target regulatory sequences, resulting in activation or repression of gene expression
3 Transcriptional Regulation of CCS Genes
Although the expression patterns and functions of genes involved in the development of the cardiac conduction system are relatively well studied, little is known about the molecular mechanisms underlying their regulation of expression. Tissue-specific gene expression often involves long-range regulatory elements, such as enhancers, which dictate the strictly time-, tissue- and dosage-dependent expression of their target genes. The identification and function of such enhancers is therefore highly relevant to fully understand the complex regulatory networks in CCS formation. However, to date only few studies have been carried out investigating in depth the regulation of genes driving CCS development.
The T-box transcription factor Tbx5 plays indispensable roles in the early patterning of the heart and CCS and is involved in limb development [15, 20, 21]. Mutations in TBX5 are associated with Holt-Oram syndrome, a developmental disorder characterized by hand-heart defects [22, 23]. Using modified bacterial artificial chromosomes (BACs), the regulatory landscape of the TBX5 locus was determined, and within this landscape, multiple cardiac-specific enhancers were identified by utilizing multiple genome-wide ChIP-seq datasets and evolutionary conservation. These enhancers were shown to recapitulate part of the TBX5 expression pattern in the heart, but interestingly, none of these fragments drove reporter expression in the limbs, suggesting that the cis-regulation of TBX5 in the heart and limbs is compartmentalized [24]. Such knowledge can highly improve the understanding of the mechanisms underlying the development of congenital heart diseases by decoupling the heart and hand phenotypes seen with Holt-Oram syndrome, thereby presenting more compartmentalized phenotypes compared to disorders caused by protein-coding mutations.
Another example of how enhancer-mediated gene expression is involved in the tight regulation of the CCS is presented by the transcriptional repressor Id2. This factor was identified by serial analysis of gene expression (SAGE) as having CCS-specific expression. Id2 is expressed throughout development in the AVB and BB and in non-CCS compartments such as the AV endocardial cushions and valves. The requirement of Id2 for ventricular CCS structure and function was demonstrated as Id2-deficient mice exhibit structural and functional conduction system abnormalities, including left bundle branch block. Id2 is cooperatively regulated by Nkx2-5 and Tbx5 in the developing ventricular conduction system by binding of both Tbx5 and Nkx2-5 to a 1052 bp fragment of the Id2 promoter. Mutation of the Tbx5 binding site within this promoter region completely abolished CCS expression whereas extracardiac expression was unaltered, illustrating the specificity of this transcriptional mechanism in the coordinated development of the ventricular conduction system [20].
The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Using a transgenic BAC approach, it was shown that the regulatory regions sufficient to recapitulate the endogenous Hcn4 expression pattern in the SAN, AVN, His bundle, bundle branches and left ventricular Purkinje fibres reside within the region covered by one bacterial artificial chromosome (BAC) of 200 kbp [25]. Using transgenic mouse assays, multiple evolutionary conserved cis-acting regulatory sequences were identified to drive Hcn4 expression in the AV conduction system. One of these regions drives reporter expression specifically in the non-chamber myocardium in a Mef2c-dependent manner. Furthermore, depletion of histone deacetylases resulted in ectopic expression of reporter activity in chamber myocardium, revealing a role for histone modifications in Mef2c-regulated enhancer-mediated expression of Hcn4 in components of the CCS [26].
More recently, Contactin-2 (Cntn2), a cell adhesion molecule critical for neuronal patterning and ion channel clustering, was described as a marker for the ventricular conduction system, with expression in the AVB, BBs and Purkinje fibres. Using a GFP-modified BAC, the boundaries of the regulatory domain involved in the control of Cntn2 expression were identified, since reporter activity of the modified BAC completely recapitulates endogenous Cntn2 expression [27]. Such knowledge facilitates in the identification of single, individual regulatory elements driving CCS development and will greatly add to our understanding of how genes involved in the complex development of CCS components are regulated.
Enhancer function is regulated by modifications of specific histone tails that mark active or poised enhancers. Active enhancers are associated with an open, accessible chromatin state, whereas poised enhancers are associated with dense, closed chromatin. Histone modifiers such as histone deactelyases (HDACs), histone methyltransferases (HMTs) and histone acetyltransferases (HATs) therefore regulate the accessibility of long-range regulatory sequences, allowing for the binding by cell type-specific transcription factors to activate transcription in a tissue-dependent manner. Specification of the AVC is regulated by Gata4, which activates AVC enhancers in synergy with Bmp2/Smad signaling to recruit HATs such as p300 [19, 28]. This leads to H3K27 acetylation, a marker of active enhancers. In contrast, in chamber myocardium, Gata4 cooperates with HDACs and chamber-specific genes Hey1 and Hey2, leading to H3K27 deacetylation and repression (Fig. 38.1b) [19].
4 Common Genomic Variants Influence CCS Function
The importance of the strict regulation of the spatial and temporal expression of CCS genes is illustrated by findings from recent genome-wide association studies, which revealed common genomic variation to be associated with conduction parameters like PR interval and QRS duration. Such variation was identified in non-coding regions flanking genes encoding ion channels like SCN5A/10A, KCNQ1 and KCNH2 and cardiac transcription factors like NKX2-5, MEIS1 and TBX3/5, indicating they might affect the function of enhancers controlling the precise regulation of these genes [29–31]. Tbx5 is broadly expressed and acts as transcriptional activator, inducing transcription of genes involved in cardiac differentiation [15, 20]. The activity domain of Tbx3 is much more restricted and confined to the developing and mature CCS, where it acts as a transcriptional repressor, thereby imposing the pacemaker phenotype on cells within its expression domain [10, 32]. Tbx3 and Tbx5 both recognize the same regulatory sequences [33], suggesting that these factors compete for binding and implicating a fine balance between activation and repression of CS genes by these factors. The precise regulation of transcription and activity of both factors is therefore crucial for proper CCS patterning, and minor changes in regulatory elements controlling the regulation of expression of these factors could thus potentially have large consequences for CCS function and development. Knowledge of the mechanisms by which such developmental genes are regulated to exert their spatio-temporal transcriptional activity is therefore crucial in the understanding of how variation identified by GWAS influences development.
5 3D Architecture Regulates Transcription
Physical enhancer-promoter contacts are a requirement for enhancer-mediated cell type-specific gene expression, and as such, the three-dimensional topology of chromatin plays an indispensable role in gene regulation by physically wiring long-range regulatory sequences with their target promoters [34]. Several protein complexes, including CTCF, cohesin and mediator, have been proposed to be involved in the organization of these contacts [35]. Furthermore, recent data suggest that such genomic structural organizers not only mediate single enhancer-promoter contacts but also mediate the organization of the genome in relatively cell-type invariant topologically associated domains (TADs) within which sequences particularly contact each other. Genes located within the same TAD exhibit greater expression correlation than genes located in distinct ones, suggesting that such domains may act as a backbone for tissue-specific regulatory contacts [36]. The recent emergence of techniques aimed at capturing the 3D conformation of genomic loci [37] therefore provides valuable tools to elucidate regulatory mechanisms on the chromatin level.
6 Regulation of Tbx3 by a Large Regulatory Domain
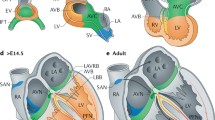
The evolutionary conserved Tbx3/5 genomic locus is one of the few genomic loci of which the 3D architecture has been studied and reveals an example of the complexity of gene regulation on the level of chromatin topology. Tbx3 and Tbx5 form an evolutionary conserved gene cluster derived from a primordial T-box gene [38] and, as mentioned above, play crucial roles in the formation and function of the cardiac conduction system. Using circular chromosome conformation capture sequencing (4C-seq), which captures all the genomic regions in close proximity to a chosen point of view [39], the 3D architecture of the Tbx3/5 locus was probed, and genomic regions contacting Tbx3 or Tbx5 were identified in different tissues (Fig. 38.2a). Interestingly, these data revealed that the regulatory landscape is in a preformed conformation that is similar in embryonic heart, brain and limb. Rather than the de novo formation of enhancer-promoter loops upon binding by cell-type relevant transcription factors to initiate transcription, the locus is in a fixed, permissive structure in which enhancer-promoter loops are pre-existing [40]. Such a permissive structure has previously been described for different loci, including the Hox and Shh gene loci. Long-range enhancer-promoter contacts in these loci were shown to be irrespective of cell type, revealing a preformed topology [36, 41]. The permissive, preformed nature of these loci was exemplified by the fact that even in the absence of a distal-acting Shh enhancer, contacts between the Shh promoter and the enhancer region still occur [41]. The benefit of such preformed regulatory landscapes is believed to lie in the ease by which tissue-specific transcription factors can utilize preformed contacts to target the gene of interest, involving only slight variations in internal contacts within an otherwise rigid and conserved structure. In agreement with this, small differences in contact profiles for the different tissue types were observed in the Tbx3/5 locus despite the fact that the domain is largely preformed, most probably caused by cell type-specific transcription factor mediated enhancer activation. Among the multiple sites contacting Tbx3 in the gene desert upstream of the gene, two evolutionary conserved enhancers have been identified that also contact each other. They are bound by cardiac-specific transcription factors Nkx2-5, Gata4, Tbx5 and Tbx3 and were shown to respond to a BMP-mediated signalling pathway to drive atrioventricular conduction system expression of Tbx3 [40].
The regulatory domains of Tbx3 and Tbx5 are physically separated. (a) Contact profiles of the Tbx3 and Tbx5 loci as determined by circular chromosome conformation capture (4C) reveal the genomic regions that physically contact Tbx3 (upper track) or Tbx5 (lower track) in mouse embryonic heart cells. Red and dark blue depict a high contact frequency, whereas light blue and grey depict a low contact frequency. The contact profiles of Tbx3 and Tbx5 hardly overlap, indicating that both genes do not share regulatory sequences and suggesting that common variants in humans upstream of TBX3 as identified by GWAS (star) can be exclusively assigned to TBX3 [40]. (b) Model of the 3D conformation of the Tbx3/Tbx5 locus. The regulatory domains of Tbx3 and Tbx5 are physically separated; however, on the protein level, both genes recognize the same binding sequences and compete with each other to activate or repress their target genes, e.g. Gja5 (Cx40). Despite the strict regulation on the chromatin level, Tbx5 also directly regulates Tbx3 by binding target sequences to activate transcription in, for example, the SAN
As mentioned before, Tbx3 and Tbx5 are expressed in overlapping patterns and have overlapping functions in CCS development. It could therefore be expected that both genes share common regulatory mechanisms on a genomic level. Studies on the transcriptional regulation of other clustered developmental genes, like the Irx and Hox clusters, revealed that regulatory sequences are not uniquely associated with single promoters, but rather are shared by multiple genes within the cluster. Such interplay between multiple enhancers coordinates the strict regulation of their expression patterns, and it has been proposed that such extensive enhancer sharing explains the conservation of the genomic organization throughout evolution [42, 43]. Interestingly, however, Tbx3 and its flanking gene desert form a loop that is physically separated from that of the neighbouring Tbx5 loop (Fig. 38.2b). Genomic regions within the Tbx3 loop solely contact Tbx3 but not Tbx5 and vice versa, indicating enhancer sharing between these evolutionary conserved clusters is unlikely to occur [40]. The strict separation of the regulatory landscapes of Tbx3 and Tbx5 is not only cell type-independent, but also evolutionary conserved between mouse and human. Recent Hi-C data in human fibroblasts [36], revealing genome-wide contact profiles, reveal a similar separation of the TBX3 and TBX5 regulatory domains with hardly any overlap of the contact profiles. This organization of the Tbx3 locus in a ~1 Mb-scale self-regulatory domain corresponds well to the previously mentioned TADs. These domains are separated by boundary regions enriched for insulator binding protein CTCF, housekeeping genes, transfer RNA and short interspersed elements, hampering interactions of sequences within one TAD with regions exceeding the domain boundaries. Regions located within the regulatory domain of Tbx3 are thus suggested to exclusively contact Tbx3 and not Tbx5 and vice versa.
7 Assigning Function to Genomic Variation
Understanding the 3D architecture of a genomic locus not only provides insight into the tight relationship between chromatin topology and the complex regulation of developmental gene expression, it could also provide valuable clues in the understanding of the role of functional variation as identified by genome-wide association studies on gene expression. Common genomic variation in the non-coding region upstream of TBX3 and TBX5 was found to influence PR interval and QRS duration in humans [29–31]. The fact that the regulatory domains of TBX3 and TBX5 are strictly separated indicates that the variation found in one of the domains can be exclusively assigned to its respective gene, facilitating our understanding of the functional effect of disease-associated variation.
A similar example of how knowledge on the chromatin conformation can increase our understanding of function of common variants is illustrated by studies on the SCN5A/SCN10A locus. SCN5A and SCN10A encode sodium channels important for conduction. GWAS implicated an intronic region in SCN10A as a major risk region for prolonged QRS duration [30]. The role of SCN10A in cardiac conduction however was not previously described, whereas mutations in the adjacent SCN5A are well established to cause several arrhythmogenic disorders, including Brugada and Long QT syndrome [44, 45]. It is therefore possible that the variation identified within the intron of SCN10A impacts the expression of SCN5A, rather than or in addition to that of SCN10A. Indeed, probing the 3D architecture with the promoters of both SCN5A and SCN10A and the site of the variation in the intron as point of view using 4C-seq revealed that this variant region contacts the SCN5A promoter and a strong enhancer downstream of SCN5A, suggesting that it might act as enhancer regulating the expression of SCN5A in the heart. Transgenic reporter assays revealed that indeed this enhancer is essential for cardiac Scn5a expression. In humans, the SNP located within the enhancer that correlates with slowed conduction is associated with lower SCN5A expression [46]. Taken together, these results provide another example of how our understanding of the 3D architecture of a genomic locus harbouring functional variation can facilitate in the assignment of function to such variations, improving our understanding of the effect of disease-associated variation.
References
Christoffels VM, Smits GJ, Kispert A, et al. Development of the pacemaker tissues of the heart. Circ Res. 2010;106:240–54.
Munshi NV. Gene regulatory networks in cardiac conduction system development. Circ Res. 2012;110:1525–37.
Stieber J, Herrmann S, Feil S, et al. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A. 2003;100:15235–40.
Baruscotti M, Bucchi A, Viscomi C, et al. Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4. Proc Natl Acad Sci U S A. 2011;108:1705–10.
Puskaric S, Schmitteckert S, Mori AD, et al. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum Mol Genet. 2010;19:4625–33.
Espinoza-Lewis RA, Yu L, He F, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev Biol. 2009;327(2):376–85.
Blaschke RJ, Hahurij ND, Kuijper S, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–8.
Espinoza-Lewis RA, Liu H, Sun C, et al. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev Biol. 2011;356:359–69.
Mommersteeg MTM, Hoogaars WMH, Prall OWJ, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–62.
Hoogaars WM, Engel A, Brons JF, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–112.
Singh R, Hoogaars WM, Barnett, et al. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol Life Sci. 2012;69:1377–89.
Boogerd KJ, Wong LYE, Christoffels VM, et al. Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of connexin 43. Cardiovasc Res. 2008;78:485–93.
Chen YH, Ishii M, Sucov HM, et al. Msx1 and Msx2 are required for endothelial-mesenchymal transformation of the atrioventricular cushions and patterning of the atrioventricular myocardium. BMC Dev Biol. 2008;8:75.
Habets PEMH, Moorman AFM, Clout DEW, et al. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–46.
Bruneau BG, Nemer G, Schmitt JP, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–21.
Lyons I, Parsons LM, Hartley L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Gene Dev. 1995;9:1654–66.
Kokubo H, Tomita-Miyagawa S, Hamada Y, et al. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134:747–55.
Singh R, Horsthuis T, Farin HF, et al. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–52.
Stefanovic S, Barnett P, van Duijvenboden K, et al. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat Commun. 2014;5:3680.
Moskowitz IP, Kim JB, Moore ML, et al. A molecular pathway including id2, tbx5, and nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–76.
Arnolds DE, Liu F, Fahrenbach JP, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–18.
Basson CT, Bachinsky DR, Lin RC, et al. Mutations in human TBX5 (corrected) cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–5.
Li QY, Newbury-Ecob RA, Terrett JA, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–9.
Smemo S, Campos LC, Moskowitz IP, et al. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet. 2012;21:3255–63.
Wu M, Peng S, Zhao Y. Inducible gene deletion in the entire cardiac conduction system using Hcn4-CreERT2 BAC transgenic mice. Genesis. 2014;52:134–40.
Vedantham V, Evangelista M, Huang Y, et al. Spatiotemporal regulation of an Hcn4 enhancer defines a role for Mef2c and HDACs in cardiac electrical patterning. Dev Biol. 2013;373:149–62.
Pallante BA, Giovannone S, Fang-Yu L, et al. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–94.
Dai YS, Markham BE. p300 Functions as a coactivator of transcription factor GATA-4. J Biol Chem. 2001;276:37178–85.
Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–9.
Sotoodehnia N, Isaacs A, de Bakker PI, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–76.
Verweij N, Mateo Leach I, van den Boogaard M, et al. Genetic determinants of P wave duration and PR segment. Circ Cardiovasc Genet. 2014;7(4):475–81.
Bakker ML, Boukens BJ, Mommersteeg MTM, et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–9.
van den Boogaard M, Wong LY, Tessadori F, et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:2519–30.
de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506.
Phillips-Cremins JE, Sauria ME, Sanyal A, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95.
Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80.
Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403.
Agulnik SI, Garvey N, Hancock S, et al. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144:249–54.
van de Werken HJ, Landan G, Holwerda SJ, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–72.
van Weerd JH, Badi I, van den Boogaard M, et al. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ Res. 2014;115(4):432–41.
Amano T, Sagai T, Tanabe H, et al. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57.
Tena JJ, Alonso ME, Calle-Mustienes E, et al. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun. 2011;2:310.
Duboule D. Vertebrate hox gene regulation: clustering and/or colinearity? Curr Opin Genet Dev. 1998;8:514–8.
Wilde AA, Brugada R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ Res. 2011;108:884–97.
Bezzina CR, Barc J, Mizusawa Y, et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45(9):1044–9.
van den Boogaard M, Smemo S, Burnicka-Turek O, et al. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J Clin Invest. 2014;124:1844–52.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution-Noncommercial 2.5 License (http://creativecommons.org/licenses/by-nc/2.5/), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited. The images or other third party material in this chapter are included in the work's Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work's Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2016 The Author(s)
About this chapter
Cite this chapter
van Weerd, J.H., Christoffels, V.M. (2016). Regulation of Vertebrate Conduction System Development. In: Nakanishi, T., Markwald, R., Baldwin, H., Keller, B., Srivastava, D., Yamagishi, H. (eds) Etiology and Morphogenesis of Congenital Heart Disease. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54628-3_38
Download citation
DOI: https://doi.org/10.1007/978-4-431-54628-3_38
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54627-6
Online ISBN: 978-4-431-54628-3
eBook Packages: MedicineMedicine (R0)