Abstract
The zebrafish (Danio rerio) has emerged in recent years as a powerful vertebrate model to study neuronal circuit development and function, thanks to its relatively small size, rapid external development and translucency. These features allow the easy application of in vivo microscopy analysis and optical perturbation of neuronal function. So far, genetic manipulation in zebrafish has been limited to the generation of constitutive loss-of-function alleles and transgenic models. CRISPR/Cas9 offers unprecedented possibilities for genomic manipulation that can be exploited to study neuronal function. In the past few years, we have successfully used CRISPR/Cas9-based technology in zebrafish to achieve two goals crucial for neuronal circuit analysis by developing two CRISPR/Cas9-based approaches that overcome previous major limitations to the study of gene and neuron functions in zebrafish. The study of gene function via tissue- or cell-specific mutagenesis remains challenging in zebrafish when the study of the function of certain loci might require tight spatiotemporal control of gene inactivation, which is particularly true in studying the function of a particular gene in post mitotic neurons, when the same gene may have had an earlier developmental function. To circumvent this limitation, we developed a simple and versatile protocol to achieve tissue-specific and temporally controlled gene disruption based on Cas9 expression under the control of the Gal4/UAS binary system (Di Donato et al. 2016). This strategy allows us to induce somatic mutations in genetically labeled cell clones or single cells and to follow them in vivo via reporter gene expression. We have also been able to target endogenous genomic loci to specifically label the great variety of neuronal cell types with reporter genes such as the transcriptional activator Gal4 (Auer et al. 2014). As a result, we can specifically target the expression of fluorescent proteins, a genetically encoded calcium indicator or optogenetic actuators in defined neuronal subpopulations.
We will present ways that these two methods can be applied to the study of the development of the nervous system in larval zebrafish.
Shahad Albadri, Flavia De Santis and Vincenzo Di Donato made equal contribution.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
CRISPR/Cas9 and Gal4/UAS Combination for Cell-Specific Gene Inactivation
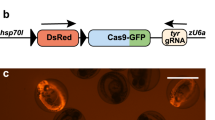
Over the last decades, the analysis of gene function has relied on mutagenesis approaches leading to the generation of loss-of-function alleles. The CRISPR/Cas9 system represents a major step forward towards achieving precise and targeted gene disruption. Being readily applicable for the creation of knockout loci in a great variety of animal models used in neuroscience studies, this technology has led to significant advances in the fields of developmental and functional neurobiology (Heidenreich and Zhang 2016). Nonetheless, constitutive gene disruption is often associated with side effects, such as compensation mechanisms and embryonic lethality, representing an important limitation on the analysis of phenotypes specific to the nervous system, since neural circuits are fully established at late stages of development. Recently, studies in worms (Shen et al. 2014), fruit flies (Port et al. 2014), mice (Platt et al. 2014) and zebrafish (Ablain et al. 2015) have pioneered the use of the CRISPR/Cas9 methodology to generate conditional gene knockouts via tissue-specific expression of cas9. This strategy takes advantage of cell type-specific promoters to control the spatiotemporal expression of the Cas9 enzyme. Importantly, one of the most common methodologies ensuring cell-specific expression of transgenes in zebrafish is the Gal4-UAS binary system (derived from yeast), in which the transcription of genes placed 3′ of an upstream activating sequence (UAS) relies on the DNA binding of the Gal4 transcriptional activator (Asakawa and Kawakami 2008). Gene- and enhancer-trap methods have been applied to establish a significant number of Gal4 transgenic lines (Davison et al. 2007; Asakawa et al. 2008; Scott and Baier 2009; Kawakami et al. 2010; Balciuniene et al. 2013), several of which are neural-specific (Scott et al. 2007; Asakawa et al. 2008). Notably, in these lines the Gal4 open reading frame (ORF) is randomly integrated in the fish genome through Tol2-based transposition, and the insertion site is not mapped; therefore, the sequence of the promoter elements driving Gal4 expression is unknown. In our work, we have developed a flexible conditional knockout strategy based on the CRISPR/Cas9 technology that combines Gal4/UAS-mediated expression of the Cas9 enzyme with a constitutive expression of sgRNAs driven by PolIII U6 promoter sequences. Our strategy does not require previous knowledge of promoter sequences to induce cas9 expression since this is provided by cell type-specific Gal4 transcription. Additionally, to enable the analysis of the phenotypes arising from Cas9-induced gene disruption, we marked the population of the cas9-expressing cells by using the viral T2A self-cleaving peptide (Provost et al. 2007), ensuring the stoichiometric synthesis of the Cas9 enzyme and the fluorescent reporter GFP from the same mRNA. To test our conditional knockout strategy, we used our vector system to target the tyrosinase (tyr) locus, coding for a key enzyme involved in melanin production (Camp and Lardelli 2001). We were able to induce eye-specific loss of pigmentation by expressing our transgene exclusively in the progenitors of the neural retina and the retinal-pigmented epithelium (RPE). For this purpose we used a transgenic line, Tg(rx2:gal4), in which the Gal4 trans-activator is specifically driven in the optic primordium by the promoter of the zebrafish retinal homeobox gene 2 (rx2; Heermann et al. 2015). This result confirmed the ability of our strategy to induce Gal4- and Cas9-mediated tissue-specific gene inactivation. Remarkably, in this first approach, GFP expression was strictly dependent on the temporal activity of the promoter driving Gal4 expression, thus restricting direct detection of potential mutant cells to a limited time window. This caveat reduces the possibility of analyzing loss-of-function phenotypes after Gal4 transactivation activity has terminated. To circumvent this issue, we proposed to use the activity of the Cre enzyme, a topoisomerase that catalyzes the site-specific recombination of DNA between loxP sites (Branda and Dymecki 2004; Pan et al. 2005), to constitutively label the population of Cas9-expressing cells. We therefore developed a construct where we substituted the GFP with a Cre reporter, enabling the analysis of gene disruption after Cas9 activity has terminated. The visualization of cre-expressing cells is commonly achieved with the use of transgenic lines carrying a cassette where a constitutive promoter drives the expression of a fluorescent reporter upon the Cre-mediated excision of a floxed stop codon. Thus, in cells carrying floxed alleles, the concomitant expression of Cas9 and Cre enzymes by a tissue-specific Gal4 promoter would ensure, respectively, double-strand breaks (DSBs) at the targeted locus as well as the recombination of the floxed locus. Notably, if the Cre-dependent expression of a reporter is constitutive after recombination, all the cells deriving from a cas9-expressing progenitor will be fluorescent, allowing long-term visualization of potentially mutated clones of cells. By using our system in retinal stem cells, we successfully disrupted the atoh7 gene, which is involved in the specification of retinal ganglion cells (RGC) in the developing retina. In this case, we could modify cell fate determination of retinal progenitor cells and generate labeled loss-of-function clones lacking the population of RGC.
Additionally, we employed our method to create genetic chimeras in which single mutant cells could be differentially tagged in a wild-type tissue. To obtain this labeling, we combined the 2C-Cas9 system with the Brainbow technology. The Tg(UAS:brainbow) line (Robles et al. 2013) carries a transgene in which the CDSs of the fluorescent proteins tdTomato, Cerulean and YFP are separated by Cre recombinase sites. In double transgenic embryos Tg(UAS:brainbow) × Tg(Tissue-specific promoter:gal4), tdTomato will be expressed in the Gal4 transactivation domain in the absence of Cre-mediated recombination. In contrast, cerulean or YFP will be transcribed if Cre recombinase is active. The expression of our transgenesis vector in these embryos provides simultaneous activity of the Cas9 and Cre enzymes. As a result, all the Gal4-positive cells that received the plasmid are potentially mutant and marked by cerulean or YFP fluorescence, whereas the population of Gal4-positive cells that do not express the construct is wild-type and labeled with the reporter tdTomato. This multicolor labeling strategy can be easily applied to neurobiology studies to induce targeted mutations in single neurons and directly compare loss-of-function and wild-type phenotypes in the same animal. To test this potential application, we targeted the genomic locus coding for the motor protein Kinesin family member 5A, a (kif5aa) (Campbell and Marlow 2013; Auer et al. 2015), whose inactivation triggers the reduction of RGC axon arbor complexity via a cell-autonomous mechanism (Auer et al. 2015). To target the kif5aa gene with the 2C-Cas9 system in single RGC, we used the Tg(isl2b:gal4) line. As expected, after injection of our construct into one-cell stage embryos derived from a cross of Tg(isl2b:gal4) and Tg(UAS:brainbow) fish, we could observe a strong decrease in total branch length in YFP- or Cerulean-expressing RGC (potentially kif5aa mutant) compared to tdTomato-fluorescent RGC (wild-type).
In conclusion, the 2C-Cas9 system represents a versatile tool to induce biallelic conditional gene inactivation. The use of the Gal4/UAS system allows the targeting of a gene of choice in any cell population. The combination of this bipartite system with simultaneous activation of Cas9 And Cre enzymes in progenitor or differentiated cells enables first, the genetic lineage tracing of mutant cells and second, the detection of cell-autonomous gene inactivation at single cell resolution. Additionally, permanent labeling of knockout cells offers the possibility of investigating gene function in adult animals, expanding the applicability of the 2C-Cas9 from neurodevelopment to maintenance and function of neural networks. Finally, because the 2C-Cas9 system is based on genetic tools available in several model organisms, this approach allows the same level of investigation in a broad range of animal models.
In addition to the use of the Crispr/Cas9 application for the generation of loss-of-function alleles, RNA guide nucleases can be used for more sophisticated genome modifications such as homologous recombination (HR) or non-homologous end joining (NHEJ)-mediated knockin. We herein provide a conceptual outline of the steps involved in the generation of knockin lines based on the Crispr/Cas9 strategy and the latest advances made in the zebrafish genome-editing field.
Crispr/Cas9-Mediated Knockin Approaches in Zebrafish
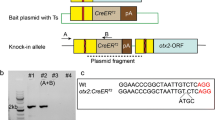
With its advantage of transparency, the zebrafish model organism rapidly emerged as a powerful experimental system for studies in genetics, developmental biology and neurobiology. The possible integration of exogenous genes into any given loci and the analysis of their function in the living animal have dramatically improved over the past few years with the development of genome editing technologies. Prior to this recent explosion in the field of knockin generation, conventional transgenic zebrafish lines were generated by Tol2-mediated transgenesis, which has successfully allowed the making of hundreds of new reporter lines essential to the study of particular gene functions in vivo (Davison et al. 2007; Asakawa et al. 2008; Scott and Baier 2009; Kawakami et al. 2010; Balciuniene et al. 2013). Bacterial artificial chromosome-based transgenesis has been and still is one of the go-to methods for making reporter lines. However, this technique comes with one major limitation: the integration of extra coding copies of hundreds of kbs. In addition, it is not known how the integration of such a large construct affects the neighboring site of insertion. More recently, the transcription activator-like effectors (TALEs) technology, a milestone in the development of zebrafish mutant and transgenic lines, has lifted the limit of loci-specific targeting. With very low off-targeting effects, TALEs were therefore the first successful genome editing method that permitted homologous-directed recombination (HDR) and NHEJ-mediated knockin in zebrafish (Bedell et al. 2012; Zu et al. 2013). Two reports (Chang et al. 2013; Hwang et al. 2013b) showed that double stranded breaks (DSB), which are simpler in design and have higher mutagenesis efficiency, could also be generated using the Crispr/Cas9 technology based on the same approach used by Bedell et al. (2012). Following these studies, Hruscha et al. (2013) achieved the integration of HA-tags into the sequence of single strand oligonucleotides flanked by two short homology arms of the targeted gene. Similarly to previously observed integration events, insertion of the sequences of interest was detected in most targeted alleles with, however, a majority of imprecise and error-prone repair mechanisms. In 2013, Zu et al. reported the first HR gene-targeting event using TALENs and a double stranded vector containing an eGFP cassette flanked by long homology arms and a germ line transmission rate of 1.5%. More recently many other laboratories have developed various methods to generate knockin alleles by HR followed by CRISPR/Cas9-induced DSB, using as donor single stranded DNA, circular or linear plasmids with short (~40 bp) or long (800–1000 bp) homology arms (Hruscha et al. 2013; Hwang et al. 2013a; Irion et al. 2014; Shin et al. 2014; He et al. 2015; Hisano et al. 2015). Although these methods were proven possible, their efficiency remains variable. To circumvent these problems, in 2014 our laboratory employed a strategy taking advantage of homologous independent repair events shown to be tenfold more active than HR events in the one-cell stage embryo (Auer and Del Bene 2014; Auer et al. 2014). The plasmid donor vector was engineered with an eGFP bait cassette and a Gal4 transcriptional transactivator cassette. Co-injected with a locus-specific sgRNA, an eGFP targeting sgRNA and cas9 nuclease mRNA, cleavage of the donor vector was generated along with the endogenous chromosomal integration site. For better readout, the injection was performed into an outcross of two transgenic lines, the first being an eGFP reporter line and the second a Tg(UAS:RFP) line. Injected embryos with a successful in-frame integration event (most probably through homologous independent repair mechanisms) therefore displayed RFP signal in cells where GFP signal was normally detected. In this system, the offspring transmission was evaluated at about 30% and increased to 40% when a selection for the RFP signal was performed after injection. The generation of such a donor vector allowed the direct assessment of the efficiency of the strategy by targeting an endogenous locus of the zebrafish genome. Targeting the transcriptional starting site of the kif5aa gene, integration of the donor vector was successfully induced and shown to be independent from the orientation of the sgRNA targeting kif5aa. In addition, no homologous sequences between the vector and the endogenous targeted site were required for the integration, allowing the re-use of the vector in combination with any given site-specific sgRNA. Using the same approach, Kimura et al. (2014) improved the strategy by adding a heat shock cassette (Hsp70) upstream of the transcription trans-activator Gal4 cassette into the donor vector, allowing its expression independently from in-frame insertion events within the transcriptional starting site of the gene of interest. To date, several new reporter lines have been generated using this strategy, providing a powerful alternative for homology-independent repair over HR-mediated integration. Key points for its success are (1) the identification of efficient sgRNAs targeting the chromosomal site of choice, for which new prescreening methods have been developed (Carrington et al. 2015; Prykhozhij et al. 2016); (2) the injection of the sgRNA mix with Cas9 nuclease mRNA over purified Cas9 protein that seems to prevent the donor plasmid insertion; and (3) further screening for the identification of founders due to the error-prone nature of junction sites between the endogenous locus and the donor vector. Hisano et al. (2015) addressed this last point by introducing 10–40 bp homology arms into the donor vector to trigger integration events mediated by HR repair mechanisms. In parallel, Li et al. (2015) developed another approach by targeting intronic regions of the gene of interest, therefore non-HR dependent. While this strategy allows keeping the integrity of the targeted coding sequence, the enriched presence of repeat sequences within the introns makes it difficult to achieve a specific targeting. Finally, the latest advance in knockin approaches is the development of traceable genome editing events that allow the easy recovery of edited alleles (Hoshijima et al. 2016) (Fig. 1).
Knockout and knockin strategies based on the Crispr/Cas9 technology in zebrafish. Schematic representation of the different methods and applications of Crispr/Cas9-mediated genome modifications. From top to bottom: (1) labeling with GFP of cas9-expressing cells potentially mutated in locus targeted by the sgRNA1 and sgRNA2 expressed with the PolIII U6 promoters. (2) Genetic labeling with Cre recombinase of cas9-expressing cells. Cre activity was revealed by the conditional expression of a fluorescent reporter protein (XFP) after removal of a stop cassette. (3) A similar strategy combined with a brainbow reporter cassette allows the visualization of cas9-expressing cells in multiple colors. (4) Genetic knockin of a Gal4 reporter transcription factor into GFP locus of preexisting transgenic lines or (5) into an endogenous genomic location (geneX). UAS upstream activating sequence
References
Ablain J, Durand EM, Yang S, Zhou Y, Zon LI (2015) A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell 32:756–764
Asakawa K, Kawakami K (2008) Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ 50:391–399
Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K (2008) Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci USA 105:1255–1260
Auer TO, Del Bene F (2014) CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods 69:142–150
Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24:142–153
Auer TO, Xiao T, Bercier V, Gebhardt C, Duroure K, Concordet J-P, Wyart C, Suster M, Kawakami K, Wittbrodt J, Baier H, Del Bene F (2015) Deletion of a kinesin I motor unmasks a mechanism of homeostatic branching control by neurotrophin-3. Elife 4:418
Balciuniene J, Nagelberg D, Walsh KT, Camerota D, Georlette D, Biemar F, Bellipanni G, Balciunas D (2013) Efficient disruption of zebrafish genes using a Gal4-containing gene trap. BMC Genomics 14:619
Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG II, Tan W, Penheiter SG, Ma AC, Leung AYH, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491:114–118
Branda CS, Dymecki SM (2004) Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6:7–28
Camp E, Lardelli M (2001) Tyrosinase gene expression in zebrafish embryos. Dev Genes Evol 211:150–153
Campbell PD, Marlow FL (2013) Temporal and tissue specific gene expression patterns of the zebrafish kinesin-1 heavy chain family, kif5s, during development. Gene Expr Patterns 13:271–279
Carrington B, Varshney GK, Burgess SM, Sood R (2015) CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucl Acids Res 43:e157–e157
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong J-W, Xi JJ (2013) Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 23:465–472
Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ (2007) Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol 304:811–824
Di Donato V, De Santis F, Auer TO, Testa N, Sánchez-Iranzo H, Mercader N, Concordet J-P, Del Bene F (2016) 2C-Cas9: a versatile tool for clonal analysis of gene function. Genome Res 26:681–692
He MD, Zhang FH, Wang HL, Wang HP, Zhu ZY, Sun YH (2015) Efficient ligase 3-dependent microhomology-mediated end joining repair of DNA double-strand breaks in zebrafish embryos. Mutat Res 780:86–96
Heermann S, Schütz L, Lemke S, Krieglstein K, Wittbrodt J (2015) Eye morphogenesis driven by epithelial flow into the optic cup facilitated by modulation of bone morphogenetic protein. Elife 4:373
Heidenreich M, Zhang F (2016) Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci 17:36–44
Hisano Y, Sakuma T, Nakade S, Ohga R, Ota S, Okamoto H, Yamamoto T, Kawahara A (2015) Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci Rep 5:8841
Hoshijima K, Jurynec MJ, Grunwald DJ (2016) Precise editing of the zebrafish genome made simple and efficient. Dev Cell 36:654–667
Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B (2013) Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140:4982–4987
Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh J-RJ (2013a) Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 8:e68708
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK (2013b) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Irion U, Krauss J, Nüsslein-Volhard C (2014) Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141:4827–4830
Kawakami K, Abe G, Asada T, Asakawa K, Fukuda R, Ito A, Lal P, Mouri N, Muto A, Suster ML, Takakubo H, Urasaki A, Wada H, Yoshida M (2010) zTrap: zebrafish gene trap and enhancer trap database. BMC Dev Biol 10:105
Kimura Y, Hisano Y, Kawahara A, Higashijima S-I (2014) Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 4:6545
Li J, Zhang BB, Ren YG, SY G, Xiang YH, JL D (2015) Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Res 25:634–637
Pan X, Wan H, Chia W, Tong Y, Gong Z (2005) Demonstration of site-directed recombination in transgenic zebrafish using the Cre/loxP system. Transgenic Res 14:217–223
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455
Port F, Chen H-M, Lee T, Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A 111:E2967–E2976
Provost E, Rhee J, Leach SD (2007) Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis 45:625–629
Prykhozhij SV, Rajan V, Berman JN (2016) A guide to computational tools and design strategies for genome editing experiments in zebrafish using CRISPR/Cas9. Zebrafish 13:70–73
Robles E, Filosa A, Baier H (2013) Precise lamination of retinal axons generates multiple parallel input pathways in the tectum. J Neurosci 33:5027–5039
Scott EK, Baier H (2009) The cellular architecture of the larval zebrafish tectum, as revealed by gal4 enhancer trap lines. Front Neural Circ 3:13
Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H (2007) Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Publ Group 4:323–326
Shen Z, Zhang X, Chai Y, Zhu Z, Yi P, Feng G, Li W, Ou G (2014) Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev Cell 30:625–636
Shin J, Chen J, Solnica-Krezel L (2014) Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development 141:3807–3818
Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S (2013) TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Meth 10:329–331
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2017 The Author(s)
About this chapter
Cite this chapter
Albadri, S., De Santis, F., Di Donato, V., Del Bene, F. (2017). CRISPR/Cas9-Mediated Knockin and Knockout in Zebrafish. In: Jaenisch, R., Zhang, F., Gage, F. (eds) Genome Editing in Neurosciences. Research and Perspectives in Neurosciences. Springer, Cham. https://doi.org/10.1007/978-3-319-60192-2_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-60192-2_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-60191-5
Online ISBN: 978-3-319-60192-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)