Abstract
In clinical neuroscience, task-based fMRI (tfMRI) is a popular method to explore the brain network activation difference between healthy controls and brain diseases like Prenatal Alcohol Exposure (PAE). Traditionally, most studies adopt the general linear model (GLM) to detect task-evoked activations. However, GLM has been demonstrated to be limited in reconstructing concurrent heterogeneous networks. In contrast, sparse representation based methods have attracted increasing attention due to the capability of automatically reconstructing concurrent brain activities. However, this data-driven strategy is still challenged in establishing accurate correspondence across individuals and characterizing group-wise consistent activation maps in a principled way. In this paper, we propose a novel multi-stage sparse coding framework to identify group-wise consistent networks in a structured method. By applying this novel framework on two groups of tfMRI data (healthy control and PAE), we can effectively identify group-wise consistent activation maps and characterize brain networks/regions affected by PAE.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
TfMRI has been widely used in clinical neuroscience to understand functional brain disorders [1]. Among all of state-of-the-art tfMRI analysis methodologies, the general linear model (GLM) is the most popular approach in detecting functional networks under specific task performance [2]. The basic idea underling GLM is that task-evoked brain activities could be discovered by subtracting the activity from a control condition [3, 4]. In common practice, experimental and control trials are performed several times and fMRI signals are averaged to increase the signal-to-noise ratio [3]. Thus task-dominant brain activities are greatly enhanced and other subtle and concurrent activities are largely overlooked. Another alternative approach is independent component analysis (ICA) [5]. However, the theoretical foundation of ICA-based methods has been challenged in recent studies [6]. Therefore, more advanced tfMRI activation detection methods are still needed.
Recently, dictionary learning and sparse representation methods have been adopted for fMRI data analysis [6, 7] and attracted a lot of attention. The basic idea is to factorize the fMRI signal matrix into an over-complete dictionary of basis and a coefficient matrix via dictionary learning algorithms [8]. Specifically, each dictionary atom represents the functional activity of a specific brain network and its corresponding coefficient vector stands for the spatial distribution of this brain network [7]. It should be noticed that the decomposed coefficient matrix naturally reveals the spatial patterns of the inferred brain networks. This novel strategy naturally accounts for the various brain networks that might be involved in concurrent functional processes [9, 10].
However, a notable challenge in current data-driven strategy is how to establish accurate network correspondence across individuals and characterize the group-wise consistent activation map in a structured method. Since each dictionary is learned in a data driven way, it is hard to establish the correspondence across subjects. To address this challenge, in this paper, we propose a novel multi-stage sparse coding framework to identify diverse group consistent brain activities and characterize the subtle cross group differences under specific task conditions. Specifically, we first concatenate all the fMRI dataset temporally and adopt dictionary learning method to identify the group-level activation maps across all the subjects. After that, we constrain spatial/temporal features in dictionary learning procedure to identify individualized temporal pattern and spatial pattern from individual fMRI data. These constrained features naturally preserve the correspondence across different subjects. Finally, a statistical mapping method is adopted to identify group-wise consistent maps. In this way, the group-wise consistent maps are identified in a structured way. By applying the proposed framework on two groups of tfMRI data (healthy control and PAE groups), we successfully identified diverse group-wise consistent brain networks for each group and specific brain networks/regions that are affected by PAE under arithmetic task.
2 Materials and Methods
2.1 Overview
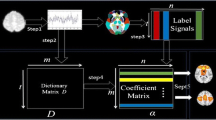
Figure 1 summarizes the computational pipeline of the multi-stage sparse coding framework. There are four major steps. First, we concatenate all the subjects’ datasets temporally to form a concatenated time*voxels data matrix (Fig. 1a) and employ the dictionary learning and sparse coding algorithms [8] to identify the group-level activation maps in the population. Then for each subject’s fMRI data, we adopt supervised dictionary learning method constraining group-level spatial patterns to learn the individualized dictionary for each subject (Fig. 1b). These individualized dictionaries are learned from individual data and thus the subject variety is better reserved. After that, for each subject, supervised dictionary learning constraining the individual dictionary is adopted to learn individualized coefficient matrix for each subject (Fig. 1c). In this way, the individualized spatial maps are reconstructed. Finally, based on the correspondence established in our method, statistical coefficient mapping method is then adopted to characterize the group-consistent activation maps for each group (Fig. 1d). Therefore, the correspondence between different subjects is preserved in the whole procedure and the group-consistent activation maps are identified in a structured method.
The computational framework of the proposed methods. (a) Concatenated sparse coding. \( t \) is the number of time point number and \( n \) is the voxel number and \( k \) is the dictionary atom number. (b) Supervised dictionary learning with spatial maps fixed. (c) Supervised dictionary learning with temporal features fixed. (d) Statistical mapping to identify group-wise consistent maps for each group.
2.2 Data Acquisition and Pre-processing
Thirty subjects participated in the arithmetic task-based fMRI experiment under IRB approval [11]. They are young adults aging from 20–26 and are from two groups: unexposed health control (16 subjects) and PAE affected ones (14 subjects). Two participants from healthy control group are abandoned due to the poor data quality. All participants were scanned in a 3T Siemens Trio scanner and 10 task blocks were alternated between a letter-matching control task and a subtraction arithmetic task. The acquisition parameters are as follows: TR = 3 s, TE = 32 ms, FA = 90, the resolution is 3.44 mm × 3.44 mm × 3 mm and the dimension is 64 × 64 × 34. The preprocessing pipeline was performed in FSL [12] including motion correction, slice time correlation, spatial smoothing, and global drift removal. The processed volumes were then registered to the standard space (MNI 152) for further analysis.
2.3 Dictionary Learning and Sparse Representation
Given the fMRI signal matrix S \( \subseteq {\mathbb{R}}^{L \times n } \), where L is the fMRI time points number and n is the voxel number, dictionary learning and sparse representation methods aim to represent each signal in S with a sparse linear combination of dictionary (D) atoms and the coefficient matrix A, i.e., S = D×A. The empirical cost function is defined as
where \( \varvec{D} \) is the dictionary, \( \ell \) is the loss function, n is the voxel number and, \( s_{i} \) is a training sample which represents the time course of a voxel. This problem of minimizing the empirical cost could be further rewritten as a matrix factorization problem with sparsity penalty:
where \( \lambda \) is a sparsity regularization parameter, \( k \) is the number of dictionary atom number and \( C \) is the set defined by the constraint to prevent D having arbitrarily large values. In order to solve this problem, we adopt the online dictionary learning and sparse coding method [8] and the algorithm pipeline is summarized in Algorithm 1 below.

2.4 Constrain Spatial Maps in Dictionary Learning
In this section, we adjust the dictionary learning procedure to constrain spatial maps in dictionary learning procedure to learn the individualized dictionary. Similar to GLM, we name each identified network as activation map. First, each group of activation map is transferred into binary vector matrix \( \varvec{V} \in \{ 0,1\}^{k \times n} \) by thresholding. Since both \( A \) and \( S \) share the same number of voxels, they have similar structures. We set all these vectors \( \varvec{V} \) as constrains in updating coefficient matrix. Specifically, if the coefficient matrix element in corresponding constrain matrix location is zero, this elements will be replaced with 0.1 (other small nonzero value is acceptable) to keep this element ‘active’. It should be noticed that the coefficient matrix is updated except that part of the elements keeps ‘active’ (nonzero). The coefficient matrix updating procedure could be represented as follows.
2.5 Constrain Temporal Features in Dictionary Learning
In our method, the dictionary is set as a fully fixed dictionary and the learning problem becomes an easy regression problem. Specifically, this dictionary learning and sparse representation problem leads to the following formulation:
where \( {\mathbf{D}}_{\text{c}} \) is the fixed individualized dictionary, \( k \) is the dictionary atom number, and A is the learned coefficient matrix from each individual fMRI data with constrained individualized dictionary in dictionary learning procedure.
2.6 Statistical Mapping
With the help of constrained features in dictionary learning procedure, the correspondences of spatial activation maps between different subjects are naturally preserved. In order to reconstruct accurate consistency maps between different groups, we hypothesize that each element in coefficient matrix is group-wisely null and a standard T-test is carried out to test the acceptance of the hypothesis. Specifically,
where \( \overline{{A_{Gx} (i,j)}} \) represents the average value of the elements in each group and \( x \) represents the patient group or control group. Specifically, the T-test acceptance threshold is set as \( p < 0.05 \). The derived T-value is further transformed to the standard z-score. In this way, each group generated a group consistent Z statistic map and each row in Z can be mapped back to brain volume standing for the spatial distribution of the dictionary atom.
3 Experimental Results
The proposed framework was applied to two groups of tfMRI data: unexposed healthy control and PAE patients. In each stage, the dictionary size is 300 and the sparsity is around 0.05 and the optimization method is stochastic approximations. Briefly, we identified 263 meaningful networks in concatenated sparse coding stage and 22 of them were affected by PAE. The detailed experimental results are reported as follows.
3.1 Identified Group-Level Activation Maps by Concatenated Sparse Coding
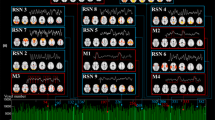
Figure 2 shows a few examples of identified group-level activation maps by concatenated sparse coding in Fig. 1a From these figures, we can see that both GLM activation map as well as common resting state networks [13] are identified, which indicates that sparse coding based methods are powerful in identifying diverse and concurrent brain activities. The quantitative measurement is shown in Table 1. The spatial similarity is defined as:
Examples of identified meaning networks by concatenated sparse coding. The first row is the template name and the second row is the template spatial map. The third row is the corresponding component network number in concatenated sparse coding. The last row is the corresponding spatial maps in concatenated sparse coding. RSN represents common resting state network in [13] and GLM result is computed from FSL feat software.
where \( {\text{X}} \) is the learned spatial network from \( A_{l} \) and \( {\text{T}} \) is the RSN template.
3.2 Learned Individualized Temporal Patterns
After concatenated sparse coding, in order to better account for subject activation variety, we constrained these identified spatial patterns in dictionary learning procedure and learned individualized temporal patterns (the method is detailed in Sect. 2.4) for each subject. Figure 3 shows two kinds of typically learned individualized temporal patterns and the correlation matrix between different subjects. Specifically, Fig. 3a shows the learned temporal patterns from constraining task-evoked group activation map (Network #175 in Fig. 2). The red line is the task design paradigm which has been convoluted with hemodynamic response function. It is interesting to see that the learned individualized temporal patterns from constraining task-evoked activation map are quite consistent and the average of these learned temporal patterns is similar to the task paradigm regressor. The correlation matrix between subjects in healthy control group is visualized in the right map in Fig. 3a and the average value is as high as 0.5. Another kind of dictionary patterns are learned from constraining resting state networks. Figure 3b shows the learned temporal patterns and correlation matrix between the healthy control group subjects with constraining resting state network (#152 in Fig. 2). The temporal patterns are quite different among different subjects and the average correlation value is as low as 0.15. From these results, we can see that the learned individualized temporal patterns are reasonable according to current neuroscience knowledge and the subtle temporal activation pattern differences among different subjects under the same task condition are recognized with the proposed framework (Fig. 4).
Identified individualized temporal patterns and correlation matrix between different subjects. (a) Identified individualized temporal patterns by constraining the same task-evoked activation map (identified in concatenated sparse coding) in dictionary learning procedure. The red line is the task paradigm pattern and the other lines are derived individualized temporal activity patterns from healthy control group subjects for the same task-evoked activation map. The right figure is the correlation matrix between different subjects. (b) Identified individualized temporal patterns by constraining resting state activation map (identified in concatenated sparse coding).
Examples of identified group-wise activation map in different groups. (a) and (b) are organized in the same fashion. The first row shows the component number and the second row shows the concatenated sparse coding results. While the third row shows the reconstructed statistical activation map in healthy control group, the last row shows the statistical activation map in PAE group. Blue circles highlight the difference between statistical maps in two groups.
3.3 Affected Activation Networks by Prenatal Alcohol Exposure
In order to identify individualized spatial activation maps, we then constrained individualized dictionary in dictionary learning procedure (detailed in Sect. 2.5) for each subject. These fixed features naturally preserve the correspondence information between subjects. After that, we adopted statistical mapping in Sect. 2.6 to generate statistical group-wise consistency maps for each group. It is easy to see that although the general spatial shapes are similar, there are subtle difference between different group statistical consistent maps which indicated the multi-stage sparse coding better captures the individual variety. Specifically, blue circles highlight the brain regions that are difference between healthy control group and PAE group. These areas includes left inferior occipital areas, left superior, right inferior parietal regions, and medial frontal gyrus which have been reported related to Prenatal Alcohol Exposure [11]. Further, it is also interesting to see that there is a clear reduction of region size in corresponding group consistency networks suggesting the similar effect of Prenatal Alcohol Exposure reported in the literature [11].
4 Conclusion
We proposed a novel multi-stage sparse coding framework for inferring group consistency maps and characterizing the subtle group response differences under specific task performance. Specifically, we combined concatenated sparse coding and supervised dictionary learning methods and statistical mapping method together to identify statistical group consistency maps in each group. This novel framework greatly overcomes the limitation of lacking correspondence between different subjects in current sparse coding based methods and provides a structured way to identify statistical group consistent maps. Experiments on healthy control and PAE tfMRI data have demonstrated the great advantage of the proposed framework in identifying meaningful diverse group consistency brain networks. In the future, we will further investigate the evaluation of subjects’ individual maps in the frame work and parameter optimization and test our framework on a variety of other tfMRI datasets.
References
Matthews, P.M., et al.: Applications of fMRI in translational medicine and clinical practice. Nat. Rev. Neurosci. 7(9), 732–744 (2006)
Fox, M.D., et al.: The human brain is intrinsically organized into dynamic, anticorrelated functional networks. PNAS 102(27), 9673–9678 (2005)
Mastrovito, D.: Interactions between resting-state and task-evoked brain activity suggest a different approach to fMRI analysis. J. Neurosci. 33(32), 12912–12914 (2013)
Friston, K.J., et al.: Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp. 2(4), 189–210 (1994)
Mckeown, M.J., et al.: Spatially independent activity patterns in functional MRI data during the stroop color-naming task. PNAS 95(3), 803–810 (1998)
Lee, K., et al.: A data-driven sparse GLM for fMRI analysis using sparse dictionary learning with MDL criterion. IEEE Trans. Med. Imaging 30(5), 1076–1089 (2009)
Lv, J., et al.: Sparse representation of whole-brain fMRI signals for identification of functional networks. Med. Image Anal. 1(20), 112–134 (2014)
Mairal, J., et al.: Online learning for matrix factorization and sparse coding. J. Mach. Learn. Res. 11, 19–60 (2010)
Pessoa, L.: Beyond brain regions: network perspective of cognition–emotion interactions. Behav. Brain Sci. 35(03), 158–159 (2012)
Anderson, M.L., Kinnison, J., Pessoa, L.: Describing functional diversity of brain regions and brain networks. Neuroimage 73, 50–58 (2013)
Santhanam, P., et al.: Effects of prenatal alcohol exposure on brain activation during an arithmetic task: an fMRI study. Alcohol. Clin. Exp. Res. 33(11), 1901–1908 (2009)
Jenkinson, M., Smith, S.: A global optimization method for robust affine registration of brain images. Med. Image Anal. 5(2), 143–156 (2001)
Smith, S.M., et al.: Correspondence of the brain’s functional architecture during activation and rest. PNAS 106(31), 13040–13045 (2009)
Acknowledgements
J. Han was supported by the National Science Foundation of China under Grant 61473231 and 61522207. X. Hu was supported by the National Science Foundation of China under grant 61473234, and the Fundamental Research Funds for the Central Universities under grant 3102014JCQ01065. T. Liu was supported by the NIH Career Award (NIH EB006878), NIH R01 DA033393, NSF CAREER Award IIS-1149260, NIH R01 AG-042599, NSF BME-1302089, and NSF BCS-1439051.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Zhao, S. et al. (2016). A Multi-stage Sparse Coding Framework to Explore the Effects of Prenatal Alcohol Exposure. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9900. Springer, Cham. https://doi.org/10.1007/978-3-319-46720-7_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-46720-7_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46719-1
Online ISBN: 978-3-319-46720-7
eBook Packages: Computer ScienceComputer Science (R0)