Abstract
Modelling provides an effective means of integrating the complementary strengths of biodiversity data derived from in situ observation versus remote sensing. The use of modelling in biodiversity change observation, or monitoring, is just one of a number of roles that modelling can play in biodiversity assessment. These roles place different levels of emphasis on explanatory versus predictive modelling, and on modelling across space alone, versus across both space and time, either past-to-present or present-to-future. One of the most challenging, yet vitally important, applications of modelling to biodiversity monitoring involves mapping change in the distribution and retention of terrestrial biodiversity. Unlike many structural and functional attributes of ecosystems, most biological entities at the species and genetic levels of biodiversity cannot be readily detected through remote sensing. Estimating change in these levels of biodiversity across large spatial extents is therefore benefiting from advances in both species-level and community-level approaches to model-based integration of in situ biological observations and remotely sensed environmental data.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
10.1 Introduction
Data on changes in the state of biodiversity on our planet come mostly from two broad sources: (1) in situ observation of organisms, or attributes of these organisms, obtained directly through application of various on-ground or in-water biological survey techniques, or through collection of museum specimens; and (2) remote sensing of biophysical characteristics of the planet’s surface detected by various satellite-borne or airborne sensors. These two sources of data have complementary strengths and weaknesses (Ferrier 2011). In situ observation provides direct information on a rich array of relevant biological entities and attributes, but the spatial coverage of such surveys is often very sparse—i.e., sampled locations are typically separated by expanses of unsurveyed land or ocean. Remote sensing, on the other hand, provides complete spatial coverage, but has limited capability to reliably detect or measure many of the biological entities or attributes of interest in biodiversity monitoring. These complementarities have, over recent decades, stimulated extensive efforts to develop change-observation methodologies that better integrate the respective strengths of in situ observation and remote sensing (Turner 2014).
While approaches to achieving this integration are many and varied, we here make an initial distinction between two broad strategies, based largely on the extent to which the biodiversity variable of interest is detectable, and therefore measurable, through remote sensing. Some types of variables are much easier to measure using remote sensing than others. For example, variables relating to ecosystem-level structural properties or functional processes—e.g., percent tree cover, canopy height, biomass, gross primary productivity (Smith et al. 2014)—tend to be more amenable to remote measurement than variables relating to species-level or genetic-level composition (Skidmore et al. 2015). Where variables, or suitable proxies, can be estimated through reasonably direct analysis or modelling of raw data from remote sensing, integration with in situ data focuses mainly on calibration and validation—i.e., using ground-based observations of the same variable as that measured remotely to calibrate (or train) the interpretation of remote data, and to test the accuracy of mapping (Baccini et al. 2007). However, in situ/remote sensing integration becomes considerably more challenging if the biological entity or attribute of interest cannot be detected readily through remote sensing—as is the case for most elements of biodiversity at the species and genetic levels.
Imagine, for example, setting out to map change in the distribution of a small forest-dwelling bird species. Unlike a variable such as percent tree cover, the presence of this species cannot be estimated directly from remote sensing. This situation demands an approach to integration that focuses less on linking in situ and remotely-sensed estimates of the same variable, and more on modelling the relationship between a variable of interest, measurable only through in situ observation, and one or more remotely mapped variables thought to be potential drivers of this variable. In this case modelling might, for example, be used to predict (or infer) change in the bird’s distribution as a function of the observed relationship between in situ data on this species and remote mapping of climate and land-cover change. In reality the measurability of variables through remote sensing forms a continuous spectrum, requiring a gradation of approaches to in situ/remote sensing integration. At one end of this spectrum in situ data are used purely to calibrate and validate estimates of a variable derived directly from remote sensing. At the other end of the spectrum, estimation of a variable of interest is made possible only by integrating in situ and remotely-sensed data through modelling, because the variable is not directly measurable through remote sensing. In this latter situation in situ data are used to calibrate and validate a model predicting the variable of interest, rather than for calibrating and validating observations of the variable itself.
Several other chapters in this book discuss applications of modelling in different fields of biodiversity monitoring—e.g. for tracking change in freshwater biodiversity in Chap. 7, in terrestrial species in Chap. 4, in genetic diversity in Chap. 5, and in ecosystem services in Chap. 3; and for adding value to remote sensing of change in Chap. 8. This chapter complements these other treatments by exploring in greater depth: (1) how the use of modelling in biodiversity monitoring relates to, and should therefore link with, the broader set of roles that modelling plays in biodiversity assessment (Sect. 10.2); and (2) the importance of matching employed modelling techniques to the particular needs of different applications in biodiversity monitoring, using as a case study the challenge of mapping change in biodiversity composition (Sect. 10.3).
10.2 Broad Roles of Modelling in Biodiversity Assessment
The use of modelling in biodiversity change observation, or monitoring, is just one of a number of roles that modelling can play in biodiversity assessment. To make better sense of this diversity of roles it is useful to first define more precisely what is actually meant by ‘modelling’ in a biodiversity context. In simple terms, a model is a set of mathematical equations (e.g., y = a + bx), or logical rules (e.g., if x > c then y = 1), that link a biodiversity variable of interest (the ‘y’ in these examples; referred to variously as the ‘dependent’, ‘response’ or ‘outcome’ variable) to one or more other variables (e.g., environmental drivers) thought to be of importance in determining, or influencing, this response (referred to variously as ‘independent’, ‘predictor’, or ‘explanatory’ variables). When publications or reports on biodiversity talk about ‘modelling’ they can be referring to either one, or sometimes both, of two quite different activities. The first of these is what we will call here ‘explanatory modelling’ (Shmueli 2010). This activity is essentially a form of data analysis, and involves using available data (observations) both for the biodiversity response variable of interest, and for the relevant predictor variables, to generate or fit a model that assesses, and describes, the relationship between these two sets of variables. In other words, known information on predictor and response variables is used to derive a model that did not exist prior to this activity. The second activity, which we here call ‘predictive modelling’ (Shmueli 2010), instead presumes that a model describing the relationship of interest is already known, as are observed or estimated values of the relevant predictor variables, and therefore combines these to predict previously unknown values of the biodiversity response variable. The model used to make such predictions can be either an ‘inductive model’ derived through data analysis, in which case the activities of data analysis and prediction are integrally linked, or a ‘deductive model’ built directly from existing expert knowledge of the relationship between response and predictor variables (Corsi et al. 2000; Overmars et al. 2007; Tuanmu and Jetz 2014).
10.2.1 Modelling Across Space Alone
Both explanatory and predictive modelling can be conducted either across geographical space, or across time, or across both space and time. The various roles played by modelling in biodiversity assessment involve different combinations of these possibilities (Figs. 10.1 and 10.2). The most basic roles are those in which modelling is conducted across space alone, at a single point in time (usually the present). Explanatory modelling of correlations, or associations, between a biodiversity response variable observed at a sample of geographical locations, and a set of predictor variables measured, or estimated, at these same locations, can help to shed light on the relative importance of different drivers in determining spatial patterns in biodiversity, and on the form (shape) of these relationships. The fitting of correlative species distribution models (SDMs) relating observations of presence, presence-absence, or abundance of a given species to multiple environmental variables (e.g., climate, terrain, soil, land-use variables) is probably the best known, and most widely applied, manifestation of such data analysis (Elith and Leathwick 2009). Other examples include statistical analyses of community-level, or ecosystem-level, attributes (e.g., species richness, functional diversity) measured at field sites distributed across different classes of land use or management (de Baan et al. 2013; Newbold et al. 2015).
Major modes of biodiversity modelling, distinguishing between explanatory and predictive modelling, and between modelling across space alone and modelling across space and time (either past-to-present or present-to-future). The shaded portion highlights those modes of most relevance to biodiversity change observation and monitoring
Explanatory modelling of drivers affecting the spatial distribution of biodiversity may be all that is required for some applications—e.g., to inform development of government policy to reduce the detrimental impact of a particular form of land use or management. However if the environmental variables used in model fitting are also mapped across an entire region of interest (e.g., as grids in a GIS) then a model derived through data analysis can, in turn, provide the foundation for prediction across geographical space (Miller et al. 2004). In the case of an SDM, this involves combining the fitted model with environmental values for each grid-cell in the region to predict occurrence within that cell, thereby producing a complete map of the predicted distribution of the species of interest. Predictive modelling of biodiversity response variables across geographical space can also be undertaken using models developed through means other than correlative data analysis. For example, the distribution of a species of interest might be predicted using a simple deductive model relating the presence or abundance of that species to mapped vegetation (or land-cover) types and/or classes of land use or management, based on expert knowledge (Stoms et al. 1992; Pearce et al. 2001; Jetz et al. 2012). Spatial prediction, whether achieved through inductive or deductive modelling, can make a vital contribution to planning and management applications requiring complete geographical mapping of biodiversity values (Guisan et al. 2013)—e.g., for the prioritisation and selection of new protected areas (Ferrier et al. 2002).
10.2.2 Modelling Across Space and Time, Present to Future
Other applications of modelling to policy development, planning and management require explanatory and/or predictive modelling to be performed not only across space, but also across time. The use of modelling to predict potential changes in biodiversity into the future, often referred to as ‘forecasts’ or ‘projections’ (Coreau et al. 2009), as a function of ongoing impacts of environmental drivers (e.g., climate and land-use change), has gained particular prominence in recent years (Pereira et al. 2010; Cook et al. 2014). Such modelling poses special challenges, as there is usually considerable uncertainty associated with the future trajectories of relevant environmental drivers, which themselves will be affected by socio-economic events and decisions that are yet to occur, and are therefore highly unpredictable. These uncertainties are often addressed through the use of scenarios—i.e., multiple plausible trajectories for environmental drivers, that account for the reality that not just one, but many, futures are possible (van Vuuren et al. 2012). Model-based biodiversity projections under plausible scenarios of change in key drivers can contribute significantly to policy agenda setting, by helping to characterise and communicate the potential magnitude of ongoing change in biodiversity, and therefore the need for action. By extending scenarios to further consider the effects of alternative policy or management interventions, such projections can also play an important role in decision support—i.e., helping policy-makers, planners and managers to choose between possible actions for addressing the problem at hand, by modelling the difference that each of these alternatives is expected to make to projected outcomes for biodiversity (Cook et al. 2014).
As for predictive modelling across geographical space, projections of biodiversity change into the future can be based on either inductive or deductive modelling (Pereira et al. 2010). When inductive models are employed for future projection, these are most often derived from correlative data analysis (i.e., explanatory modelling) of relationships between biodiversity and environmental drivers observed across space alone, rather than across time. Using such models to project changes across time involves space-for-time substitution. This assumes that the correlation observed across space between a given biological response variable, and one or more environmental variables (e.g., between the presence of a species, and climate and land use), will also hold across time, and can therefore be used to predict future changes in this response variable as a function of changing environmental conditions. While there is often little choice but to rely on space-for-time substitution for projecting future change in biodiversity, questions are increasingly being raised and examined around the robustness of this approach (Bonthoux et al. 2013; Araujo and Peterson 2012; Blois et al. 2013).
10.2.3 Modelling Across Space and Time, Past to Present
Modelling change in biodiversity across time is not limited to future projection, but is also crucially important for observing and analysing change in biodiversity that has already occurred (past to present). Modelling plays two broad roles in biodiversity change observation and monitoring, aligned directly with the distinction between explanatory modelling and predictive modelling introduced above. Where changes both in a biodiversity response of interest, and in relevant environmental drivers, are observed over both space and time, explanatory modelling of driver-response correlations can be taken to a level of rigour beyond that of modelling based on observations from across space alone (Kery et al. 2013). In addition to direct provision of stronger policy-relevant evidence for the impact of drivers on biodiversity, explanatory modelling based on temporal observations is also vital to achieving more effective integration of biodiversity monitoring (past to present) and projection (present to future). Inductive models derived through analysis of observed changes in biodiversity and environmental drivers over time are likely to provide a stronger foundation for projecting future change than projections based purely on space-for-time substitution (Santika et al. 2014). This is because models fitted to temporal data have potential to better distinguish actual drivers of change from environmental variables simply exhibiting spatial autocorrelation with these drivers, and to better account for the effects of dynamic processes that may be difficult to detect and describe based on spatial data alone (e.g., the phenotypic plasticity of species in the face of environmental change). Even more importantly, using explanatory modelling to analyse future observations generated by ongoing biological and environmental monitoring initiatives offers a powerful means of testing projections made over the same time period, thereby informing adaptive refinement of models underpinning policy and decision-making into the future (Ferrier 2012; Rapacciuolo et al. 2014).
The second major role that modelling plays in relation to biodiversity change observation and monitoring is predictive, rather than explanatory, in nature. Rather than projecting potential changes in biodiversity into the future (as described in Sect. 10.2.2), model-based prediction is used here to help fill spatial and temporal gaps in the coverage of direct observations of biodiversity change past-to-present. As noted earlier, many biological entities or attributes of interest from a biodiversity monitoring perspective can be detected only through in situ observation. Locations at which changes in these variables are measured directly therefore tend to be distributed very sparsely, and often unevenly, across the planet’s surface. In contrast, changes in environmental drivers—e.g. climate, land use—are often more amenable to detection through remote sensing, and therefore potentially mappable across large geographical extents. For applications in policy, planning or management that require complete geographical coverage of information on biodiversity change, predictive modelling can play a valuable role in translating mapped changes in key drivers, generated through remote sensing, into expected changes in biodiversity (Lung et al. 2012; Soberon and Peterson 2009). The models underpinning such translation can, again, be either deductive or inductive, with the latter derived from explanatory modelling of biological and environmental data distributed either across space alone (and therefore constituting another form of space-for-time substitution), or across both space and time.
The remainder of this chapter explores, in greater depth, this last role of modelling in biodiversity monitoring—i.e., the use of predictive modelling to help map past-to-present changes in the distribution of biodiversity across large spatial extents.
10.3 A Key Modelling Challenge: Mapping Change in the Distribution and Retention of Terrestrial Biodiversity
Unlike many structural and functional attributes at the ecosystem level, most biological entities at the species and genetic levels of biodiversity cannot be readily detected through remote sensing. Notable exceptions include the emerging use of very high spatial resolution imagery to identify individual organisms of certain large-bodied, and conspicuous, animal species (e.g., penguins; Fretwell et al. 2012), and the use of hyperspectral sensors to detect variation in plant species composition in the top layer of vegetation communities (Leutner et al. 2012). These developments offer considerable potential for direct derivation of spatially-complete mapping of temporal change from remote sensing, for at least a subset of biological entities. However this still leaves a very large proportion of our planet’s biological diversity that is effectively invisible to satellite-borne remote sensing, both at the species level and, even more so, at the genetic level. In situ monitoring of change in these components of diversity at selected locations may provide all the information that is needed for some applications—e.g., for monitoring the performance of local-scale management actions (Lindenmayer et al. 2012). Estimating change across large spatial extents—e.g., across a whole ecoregion, country or continent, or across the entire planet—poses a much greater challenge for in situ monitoring, particularly if these changes need to be mapped at relatively fine spatial resolution across the entire extent of interest (Ferrier 2011; Jetz et al. 2012; Pereira and Cooper 2006). We here explore how various modelling approaches can be used to help address this challenge, by integrating the respective strengths of data generated through in situ and remote sensing observation techniques.
Our focus is mostly on the terrestrial realm, although many of the modelling approaches discussed below are also applicable in freshwater and marine systems. We first consider ‘species-level approaches’ that model and map changes in the distribution of individual species, and then move on to examine ‘community-level approaches’ that focus instead on modelling and mapping changes in the distribution and retention of biological diversity within whole communities, without providing explicit information on the individual species comprising this diversity.
10.3.1 Species-Level Approaches
Interest in techniques for modelling, and thereby mapping, distributions of individual species as a function of remotely mapped environmental variables has grown rapidly over the past 30 years. Particularly strong attention has been directed towards correlative species distribution modelling (SDM) which uses statistical model-fitting, or machine learning, to derive explanatory models linking in situ observations of species occurrence to environmental predictors (Elith and Leathwick 2009). This largely inductive approach has been complemented, to a lesser extent, by deductive modelling of distributions based on expert knowledge of the environmental or habitat requirements of species (Jetz et al. 2012), or by more mechanistic modelling based on independently acquired evidence of ecophysiological limits or understanding of other relevant ecological factors and processes shaping species distributions (Kearney et al. 2010).
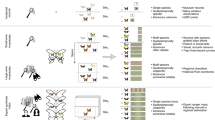
While the scientific literature on species distribution modelling is now very extensive (Guisan et al. 2013; Ahmed et al. 2015), a large proportion of these studies have focused on using such modelling to predictively map distributions across space alone, or to project potential changes in distribution into the future under alternative global-change scenarios. The use of this modelling paradigm in biodiversity monitoring—i.e., to help map past-to-present changes in species distributions—is surprisingly rare relative to these other applications. A number of options are nevertheless available for making effective use of species distribution modelling in monitoring (Fig. 10.3). To simplify the explanation of these options we will here focus on just two of the main drivers of ongoing changes in biological distributions—i.e., habitat loss or degradation (linked to changes in land cover and use) and climate change—both of which are amenable to spatially-complete change detection and mapping through remote sensing. If remotely-sensed variables relating to habitat loss or degradation are included as predictors in explanatory models fitted inductively to species occurrences observed across space alone, then such models can be used to predictively map distributional changes as a function of observed change in these variables, through simple space-for-time substitution (Lung et al. 2012). Alternatively, deductive modelling based on expert knowledge of the association between a given species and classes of land cover, or broad habitat type, can be used to predict changes in the distribution of that species as a function of remote mapping of these classes over time (Jetz et al. 2007, 2012). Considerable scope also exists to combine inductive and deductive modelling approaches in this context—e.g., by using inductive species distribution modelling, and available occurrence records, to map the ‘natural’ or ‘original’ distribution of a species as a function of mapped abiotic environmental variables (climate, terrain, soils etc.), and then using remote sensing and simple deduction to map changes over time in the portions of this distribution lost through habitat transformation (Barrows et al. 2008; Rios-Munoz and Navarro-Siguenza 2009).
Using species distribution modelling to predictively map changes in the distribution of species in response to remotely observed changes in climate is rather more challenging than for changes in land use or cover. There is potential to again employ space-for-time substitution for this purpose, by using explanatory models describing relationships between species occurrence and climate across space to predict changes in distribution across time as a function of observed changes in climate, mapped either directly from remote sensing, or through model-based integration of remotely-sensed and in situ climate observations. Many unanswered questions remain, however, regarding the transferability of climatic associations of species between space and time (Araujo and Peterson 2012). The impact of a given change in climate over time—e.g., a 0.5 °C increase in mean annual temperature—on the occurrence of a given species at a particular location may be substantially less (or in some cases more) than that observed over space, due to complicating factors such as time lags in response, capacity for phenotypic plasticity, genetic adaptation, and biological interactions.
These difficulties point to the desirability of, wherever possible, fitting explanatory models relating species’ occurrence to climate (and, for that matter, to land use or cover or abiotic variables such as soil type) using observations gathered across both time and space, rather than across space alone. Rapidly growing interest is now being directed towards extending standard techniques for species distribution modelling to more effectively consider the temporal dimension of observations (Kharouba et al. 2009; Porzig et al. 2014)—e.g., through the use of dynamic occupancy modelling (Kery et al. 2013; Tingley and Beissinger 2009). In an ideal world the fitting of explanatory models to biological and environmental observations obtained over time at a sample of locations, and the use of these fitted models to predictively map changes in biological distributions across an entire region of interest, would occur in parallel (as depicted in Fig. 10.3).
The process described so far is focused on predicting past-to-present change in the occurrence of a given species at a given location (e.g., grid cell), and thereby mapping change across all locations within a region of interest. For some applications this raw spatio-temporal information may need to be subjected to further aggregation or synthesis to address questions regarding, for example, changes in the overall state of a species, or of a whole group of species, within a given region. The most straightforward approach to deriving such aggregate measures is through simple summation, or averaging, of the predicted occurrence of a species across all locations (grid cells) in the region and, in turn, across all species in the group of interest. However it is worth noting in passing that other options exist for incorporating additional factors into this process of aggregation and synthesis—e.g., the use of metapopulation modelling to consider the effects of spatial configuration of predicted occurrence on the overall persistence of a species (Drielsma and Ferrier 2009), or the incorporation of information on phylogenetic relationships or functional traits into aggregate measures of the state of biodiversity across multiple species (Fenker et al. 2014).
10.3.2 Community-Level Approaches
In the species-level approaches discussed above, modelling is used to predictively map changes in the distribution of individual species. We now turn our attention to so-called ‘community-level approaches’ to modelling, and thereby mapping, changes in the distribution and retention of biodiversity within whole communities, without providing explicit information on the individual (named) species comprising this diversity. These approaches have particular utility in situations where the number of species in a biological group of interest is so high, and/or the average amount of information available for each of these species is so low, that species-level approaches start to lose tractability—e.g., for arthropods or plants in tropical forests. To appreciate the role that such approaches can play in mapping biodiversity change, let us start with a relatively basic challenge—i.e., estimating the loss (or, conversely, retention) of biological diversity at a single location (grid cell) as a function of remotely-sensed habitat loss or degradation. If remote sensing is used to classify the natural habitat within each grid cell in a region as being either intact or removed (Hansen et al. 2013), then simple deduction may be all that is required to predict the impact of this state on local biodiversity within that cell—i.e., it can be assumed that most of the species that were dependent on this habitat will no longer occur at this particular location. Alternatively, remote sensing can be used to classify locations into multiple classes of land use or habitat condition/intactness (Martinez and Mollicone 2012). These classes are expected to have varying levels of impact on local biodiversity. Prediction of these impacts should ideally be based on explanatory modelling of biological data gathered from the different classes, either across space alone (Souza et al. 2015) or, preferably, across both space and time (Casner et al. 2014). A particularly noteworthy example of this application of explanatory modelling is the PREDICTS initiative, which has undertaken an extensive meta-analysis of land-use impacts on local biodiversity (change in species richness) based on data for 27,000 species at over 11,000 sites globally (Newbold et al. 2015).
Linking explanatory models such as this to remotely-sensed land-use change opens up considerable potential to predictively map past-to-present change in local biodiversity across all grid cells in a region, or even across the entire planet. Change in local biodiversity is, however, not the only aspect of change that needs to be considered by community-level approaches to modelling biodiversity change. The total diversity—e.g., of species—occurring on our planet is a function not just of the number of species occurring at individual locations (alpha diversity), but also of differences in the composition of species between these locations (beta diversity) (McGill et al. 2015). To properly interpret the impacts of habitat loss (and, in turn, climate change) on retention of overall biodiversity it is therefore highly desirable to factor beta diversity into any model-based interpretation of remotely-sensed environmental change. Two broad strategies are available for achieving this, one using discrete classes to represent spatial pattern in beta diversity, and the other accounting for beta diversity through modelling of continuous patterns of spatial turnover in species composition (Ferrier 2011).
10.3.2.1 Discrete Community-Level Approaches
Many different types of discrete classification of communities can be employed in this context (Ferrier et al. 2009). The only real constraint is that the relevant classes are mapped across the entire region of interest, and that these classes provide a reasonable representation of major spatial patterns expected in the distribution of biodiversity in the absence of habitat loss or degradation. The last part of this constraint is particularly important. If the effects of habitat degradation are reflected in the classification itself (e.g., an area of rainforest cleared for domestic grazing is treated as a grassland rather than a forest) then it ceases to provide a logical basis for incorporating consideration of beta diversity into the interpretation of remotely-sensed environmental change. Mapped ecoregions may serve this purpose well at coarser spatial scales (Giam et al. 2011), as may mapping of the ‘natural’ extent (prior to anthropogenic alteration) of vegetation communities at finer scales (Keith et al. 2009).
With recent advances in the availability and resolution of abiotic environmental layers globally (for climate, terrain, soils etc.) another option growing in popularity is to derive environmental classes by integrating these layers—either by generating all unique combinations of expert-defined categories for each environmental variable (Ferrier and Watson 1997; Sayre et al. 2014), or through some form of automated numerical classification (Mackey et al. 2008). If sufficient biological data are available—i.e., in situ records for multiple species, well distributed across the region of interest—then various community-level modelling techniques can also be used to automatically derive and map environmental classes that best fit observed biological patterns (Ferrier and Guisan 2006).
Assuming that a mapped classification has been generated using one of the above approaches, this can be combined with remote mapping of habitat loss or degradation to estimate change in the retention of biodiversity. Where remote sensing yields a binary habitat versus no-habitat measure for each grid cell, then the changing state of a given class (e.g., an ecoregion) can be most simply expressed as the proportion of cells in that class with intact habitat. If remote sensing instead yields multiple levels of habitat condition/intactness—e.g., land-use classes translated into proportional losses of local species richness using results from the PREDICTS meta-analysis (described above)—then weighted averaging of these levels across all cells in a class can be used to derive an effective proportion of habitat remaining in that class (Scholes and Biggs 2005; Pereira and Daily 2006). In some cases this effective proportion is further adjusted to account for the effects of the spatial configuration of habitat—e.g., a cell with a given condition value located within a small isolated habitat fragment is assigned less weight than a cell of the same value located within a large well-connected area of habitat (Drielsma et al. 2014; Ferrier and Drielsma 2010).
Estimation of the proportion, or effective proportion, of habitat remaining in a class can be further used to predict the proportion of species, originally occurring within that class, that are expected to persist if this proportion of habitat is retained over the longer term. Such prediction is most commonly undertaken using some form of species-area relationship (SAR) (Ferrier 2002; Pereira and Daily 2006). SAR-based approaches typically assume that all classes are equally rich in species, and treat each mapped class (e.g., an ecoregion) as if it is a closed system—i.e., it is assumed that the species occurring within this class do not also occur in any of the other classes. The overall proportion of species predicted to be retained within an entire region of interest is therefore calculated as a simple average of the predicted proportions of species retained when the SAR is applied separately to each of the classes within the region (Faith et al. 2008; Proenca and Pereira 2013). Where estimates are available of the relative species richness of classes, and of the level of overlap in species composition between classes (e.g., the proportion of species occurring in ecoregion m that also occur in ecoregion n) then techniques exist for incorporating this information directly into SAR-based prediction of the overall proportion of species retained in a region as a function of remotely-sensed proportions, or effective proportions, of habitat retained in each class (Turak et al. 2011; Leathwick et al. 2010; Faith et al. 2008).
10.3.2.2 Continuous Community-Level Approaches
In the discrete community-level approaches described above, each location (e.g., grid cell) in the region of interest is viewed as belonging to a discrete class of locations that are assumed to be equally similar to one another, and equally different from locations in other classes, in the species they support. Real-world patterns of spatial change, or turnover, in species composition are, however, often more complex than can be effectively represented by a discrete classification with hard boundaries between mapped classes. Continuous community-level approaches attempt to address this reality by treating the composition of species occurring at each individual location as being unique, and the proportional overlap, or conversely distinctiveness, in composition between this location and any other given location within the region of interest as varying in a continuous manner (Ferrier et al. 2009).
One approach to applying this continuous community-level perspective to predictive mapping of change in biodiversity, as a function of remotely-sensed changes in habitat and/or climate, is through the use of generalised dissimilarity modelling (GDM) (Ferrier et al. 2007). GDM employs in situ occurrence records for all species in a given biological group (e.g., all plants, reptiles, or land snails) to fit a non-linear statistical model relating the dissimilarity in species composition observed between two locations to environmental differences based on remotely-mapped predictors (climate, terrain, soil etc.). Models fitted with GDM effectively weight and scale these environmental variables, thereby transforming multidimensional environmental space in such a way that distances within this transformed space match observed compositional dissimilarities as closely as possible. Using fitted GDM models to interpret remotely-sensed change in the distribution and condition of habitat can be achieved in various ways, but one of the most straightforward solutions is an extension of the SAR-based approach described above for the discrete community-level situation. In this extended approach the proportion, or effective proportion, of habitat remaining is estimated separately for each individual grid-cell within a region. This is calculated as a weighted average of habitat condition in all cells environmentally similar to the cell of interest, with each cell weighted by the level of similarity predicted by the fitted GDM. SAR-based estimates of the proportion of species retained relative to each cell can then be aggregated into an overall estimate of the proportion of species retained within the region as a whole (or within any required subset of this) factoring in GDM-predicted compositional dissimilarities between these cells (Ferrier et al. 2004; Allnutt et al. 2008).
Because continuous community-level approaches, such as GDM, incorporate abiotic environmental variables directly into the modelling of beta-diversity patterns, this opens up potential to further predict changes in the distribution and retention of biodiversity as a function of remotely-observed changes in climate. This can be achieved by invoking space-for-time substitution in a similar manner to that described earlier for species distribution modelling (Fitzpatrick et al. 2011; Prober et al. 2012). However it should be noted that employing space-for-time substitution in community-level approaches is also affected by many of the same complicating factors identified for species-level applications—e.g., time lags in response, capacity for phenotypic plasticity, genetic adaptation, and biological interactions (Blois et al. 2013). This again points to the desirability of fitting explanatory models relating patterns of biological distribution (in this case, turnover in species composition) to climate and habitat using observations gathered across both time and space, rather than across space alone. As for species distribution modelling, interest is now growing in extending existing community-level modelling approaches to more effectively consider the temporal dimension of biological observations.
10.4 Conclusion
As outlined in this chapter, modelling can play a crucial role in biodiversity monitoring by enabling more effective integration of in situ biological data with remotely-observed changes in key environmental drivers. This integration can involve both explanatory modelling—i.e., assessing and describing the effect of drivers on biodiversity through analysis of relationships between observed changes in biological and environmental data; and predictive modelling—i.e., using modelled relationships to predictively map change in biodiversity across whole regions as a function of remotely-sensed environmental change.
The most significant challenge now facing applications of modelling to biodiversity monitoring is to reduce reliance on models fitted to in situ biological observations gathered across space alone by making more extensive and effective use of observations from across both space and time. Recent escalation of interest in, and uptake of, citizen science initiatives (see Chap. 9) for collecting large quantities of spatially- and temporally-explicit biological observations offers considerable potential in this regard. In many cases incorporating data generated by such initiatives into biodiversity modelling will require extension of existing modelling techniques, or development of whole new techniques (Bird et al. 2014; Isaac et al. 2014; van Strien et al. 2013). These advances are likely to significantly strengthen links between explanatory and predictive modelling within the context of biodiversity monitoring. They are also likely to help strengthen links with applications of modelling to the projection of future biodiversity outcomes, by providing a more rigorous foundation both for fitting models employed in such projections, and for ongoing testing of the accuracy of these projections.
References
Ahmed, S. E., McInerny, G., O’Hara, K., Harper, R., Salido, L., Emmott, S., et al. (2015). Scientists and software—surveying the species distribution modelling community. Diversity and Distributions, 21, 258–267.
Allnutt, T., Ferrier, S., Manion, G., Powell, G., Ricketts, T., Fisher, B., et al. (2008). A method for quantifying biodiversity loss and its application to a 50-year record of deforestation across Madagascar. Conservation Letters, 1, 173–181.
Araujo, M. B., & Peterson, A. T. (2012). Uses and misuses of bioclimatic envelope modeling. Ecology, 93, 1527–1539.
Baccini, A., Friedl, M. A., Woodcock, C. E., & Zhu, Z. (2007). Scaling field data to calibrate and validate moderate spatial resolution remote sensing models. Photogrammetric Engineering and Remote Sensing, 73, 945–954.
Barrows, C. W., Preston, K. L., Rotenberry, J. T., & Allen, M. F. (2008). Using occurrence records to model historic distributions and estimate habitat losses for two psammophilic lizards. Biological Conservation, 141, 1885–1893.
Bird, T. J., Bates, A. E., Lefcheck, J. S., Hill, N. A., Thomson, R. J., Edgar, G. J., et al. (2014). Statistical solutions for error and bias in global citizen science datasets. Biological Conservation, 173, 144–154.
Blois, J. L., Williams, J. W., Fitzpatrick, M. C., Jackson, S. T., & Ferrier, S. (2013). Space can substitute for time in predicting climate-change effects on biodiversity. Proceedings of the National Academy of Sciences of the United States of America, 110, 9374–9379.
Bonthoux, S., Barnagaud, J. Y., Goulard, M., & Balent, G. (2013). Contrasting spatial and temporal responses of bird communities to landscape changes. Oecologia, 172, 563–574.
Casner, K. L., Forister, M. L., O’Brien, J. M., Thorne, J., Waetjen, D., & Shapiro, A. M. (2014). Contribution of urban expansion and a changing climate to decline of a butterfly fauna. Conservation Biology, 28, 773–782.
Cook, C. N., Inayatullah, S., Burgman, M. A., Sutherland, W. J., & Wintle, B. A. (2014). Strategic foresight: How planning for the unpredictable can improve environmental decision-making. Trends in Ecology & Evolution, 29, 531–541.
Coreau, A., Pinay, G., Thompson, J. D., Cheptou, P. O., & Mermet, L. (2009). The rise of research on futures in ecology: Rebalancing scenarios and predictions. Ecology Letters, 12, 1277–1286.
Corsi, F., de Leeuw, J., & Skidmore, A. (2000) Modelling species distribution with GIS. In L. Boitani & T. Fuller (Eds.), Research techniques in animal ecology (pp. 389–413). Columbia University Press.
de Baan, L., Alkemade, R., & Koellner, T. (2013). Land use impacts on biodiversity in LCA: A global approach. International Journal of Life Cycle Assessment, 18, 1216–1230.
Drielsma, M., & Ferrier, S. (2009). Rapid evaluation of metapopulation persistence in highly variegated landscapes. Biological Conservation, 142, 529–540.
Drielsma, M., Ferrier, S., Howling, G., Manion, G., Taylor, S., & Love, J. (2014). The biodiversity forecasting toolkit: Answering the ‘how much’, ‘what’, and ‘where’ of planning for biodiversity persistence. Ecological Modelling, 274, 80–91.
Elith, J., & Leathwick, J. R. (2009). Species distribution models: Ecological explanation and prediction across space and time. Annual Review of Ecology Evolution and Systematics, 40, 677–697.
Faith, D., Ferrier, S., & Williams, K. (2008). Getting biodiversity intactness indices right: Ensuring that ‘biodiversity’ reflects ‘diversity’. Global Change Biology, 14, 207–217.
Fenker, J., Tedeschi, L. G., Pyron, R. A., & Nogueira, C. D. (2014). Phylogenetic diversity, habitat loss and conservation in South American pitvipers (Crotalinae: Bothrops and Bothrocophias). Diversity and Distributions, 20, 1108–1119.
Ferrier, S. (2002). Mapping spatial pattern in biodiversity for regional conservation planning: Where to from here? Systematic Biology, 51, 331–363.
Ferrier, S. (2011). Extracting more value from biodiversity change observations through integrated modeling. BioScience, 61, 96–97.
Ferrier, S. (2012). Big-picture assessment of biodiversity change: Scaling up monitoring without selling out on scientific rigour. In D. Lindenmayer & P. Gibbons (Eds.), Biodiversity monitoring in Australia (pp. 63–70). Canberra: CSIRO Publishing.
Ferrier, S., & Drielsma, M. (2010). Synthesis of pattern and process in biodiversity conservation assessment: A flexible whole-landscape modelling framework. Diversity and Distributions, 16, 386–402.
Ferrier, S., Faith, D., Arponen, A., & Drielsma, M. (2009) Community-level approaches to spatial conservation prioritization. In A. Moilanen, H. Possingham & K. Wilson (Eds.), Spatial conservation prioritization: Quantitative methods and computational tools. Oxford University Press.
Ferrier, S., & Guisan, A. (2006). Spatial modelling of biodiversity at the community level. Journal of Applied Ecology, 43, 393–404.
Ferrier, S., Manion, G., Elith, J., & Richardson, K. (2007). Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Diversity and Distributions, 13, 252–264.
Ferrier, S., Powell, G., Richardson, K., Manion, G., Overton, J., Allnutt, T., et al. (2004). Mapping more of terrestrial biodiversity for global conservation assessment. BioScience, 54, 1101–1109.
Ferrier, S., & Watson, G. (1997) An evaluation of the effectiveness of environmental surrogates and modelling techniques in predicting the distribution of biological diversity. Canberra: Environment Australia. http://www.environment.gov.au/archive/biodiversity/publications/technical/surrogates/index.html
Ferrier, S., Watson, G., Pearce, J., & Drielsma, M. (2002). Extended statistical approaches to modelling spatial pattern in biodiversity in northeast New South Wales. I. Species-level modelling. Biodiversity and Conservation, 11, 2275–2307.
Fitzpatrick, M. C., Sanders, N. J., Ferrier, S., Longino, J. T., Weiser, M. D., & Dunn, R. (2011). Forecasting the future of biodiversity: A test of single- and multi-species models for ants in North America. Ecography, 34, 836–847.
Fretwell, P. T., LaRue, M. A., Morin, P., Kooyman, G. L., Wienecke, B., Ratcliffe, N., et al. (2012). An emperor penguin population estimate: The first global, synoptic survey of a species from space. PLoS ONE, 7, 11.
Giam, X. L., Sodhi, N. S., Brook, B. W., Tan, H. T. W., & Bradshaw, C. J. A. (2011). Relative need for conservation assessments of vascular plant species among ecoregions. Journal of Biogeography, 38, 55–68.
Guisan, A., Tingley, R., Baumgartner, J. B., Naujokaitis-Lewis, I., Sutcliffe, P. R., Tulloch, A. I. T., et al. (2013). Predicting species distributions for conservation decisions. Ecology Letters, 16, 1424–1435.
Hansen, M. C., Potapov, P. V., Moore, R., Hancher, M., Turubanova, S. A., Tyukavina, A., et al. (2013). High-resolution global maps of 21st-Century forest cover change. Science, 342, 850–853.
Isaac, N. J. B., van Strien, A. J., August, T. A., de Zeeuw, M. P., & Roy, D. B. (2014). Statistics for citizen science: Extracting signals of change from noisy ecological data. Methods in Ecology and Evolution, 5, 1052–1060.
Jetz, W., McPherson, J. M., & Guralnick, R. P. (2012). Integrating biodiversity distribution knowledge: Toward a global map of life. Trends in Ecology & Evolution, 27, 151–159.
Jetz, W., Wilcove, D. S., & Dobson, A. P. (2007). Projected impacts of climate and land-use change on the global diversity of birds. PLoS Biology, 5, 1211–1219.
Kearney, M. R., Wintle, B. A., & Porter, W. P. (2010). Correlative and mechanistic models of species distribution provide congruent forecasts under climate change. Conservation Letters, 3, 203–213.
Keith, D. A., Orscheg, C., Simpson, C. C., Clarke, P. J., Hughes, L., Kennelly, S. J., et al. (2009). A new approach and case study for estimating extent and rates of habitat loss for ecological communities. Biological Conservation, 142, 1469–1479.
Kery, M., Guillera-Arroita, G., & Lahoz-Monfort, J. J. (2013). Analysing and mapping species range dynamics using occupancy models. Journal of Biogeography, 40, 1463–1474.
Kharouba, H. M., Algar, A. C., & Kerr, J. T. (2009). Historically calibrated predictions of butterfly species’ range shift using global change as a pseudo-experiment. Ecology, 90, 2213–2222.
Leathwick, J., Moilanen, A., Ferrier, S., & Julian, K. (2010). Complementarity-based conservation prioritization using a community classification, and its application to riverine ecosystems. Biological Conservation, 143, 984–991.
Leutner, B. F., Reineking, B., Muller, J., Bachmann, M., Beierkuhnlein, C., Dech, S., et al. (2012). Modelling forest alpha-diversity and floristic composition—on the added value of LiDAR plus hyperspectral remote sensing. Remote Sensing, 4, 2818–2845.
Lindenmayer, D. B., Gibbons, P., Bourke, M., Burgman, M., Dickman, C. R., Ferrier, S., et al. (2012). Improving biodiversity monitoring. Austral Ecology, 37, 285–294.
Lung, T., Peters, M. K., Farwig, N., Bohning-Gaese, K., & Schaab, G. (2012). Combining long-term land cover time series and field observations for spatially explicit predictions on changes in tropical forest biodiversity. International Journal of Remote Sensing, 33, 13–40.
Mackey, B. G., Berry, S. L., & Brown, T. (2008). Reconciling approaches to biogeographical regionalization: A systematic and generic framework examined with a case study of the Australian continent. Journal of Biogeography, 35, 213–229.
Martinez, S., & Mollicone, D. (2012). From land cover to land use: A methodology to assess land use from remote sensing data. Remote Sensing, 4, 1024–1045.
McGill, B. J., Dornelas, M., Gotelli, N. J., & Magurran, A. E. (2015). Fifteen forms of biodiversity trend in the Anthropocene. Trends in Ecology & Evolution, 30, 104–113.
Miller, J. R., Turner, M. G., Smithwick, E. A. H., Dent, C. L., & Stanley, E. H. (2004). Spatial extrapolation: The science of predicting ecological patterns and processes. BioScience, 54, 310–320.
Newbold, T., Hudson, L. N., Hill, S. L. L., Contu, S., Lysenko, I., & Senior, R. A. et al. (2015) Global effects of land use on local terrestrial biodiversity. Nature, 520, 45–50.
Overmars, K. P., de Groot, W. T., & Huigen, M. G. A. (2007). Comparing inductive and deductive modeling of land use decisions: Principles, a model and an illustration from the Philippines. Human Ecology, 35, 439–452.
Pearce, J., Cherry, K., Drielsma, M., Ferrier, S., & Whish, G. (2001). Incorporating expert opinion and fine-scale vegetation mapping into statistical models of faunal distribution. Journal of Applied Ecology, 38, 412–424.
Pereira, H. M., & Cooper, H. D. (2006). Towards the global monitoring of biodiversity change. Trends in Ecology & Evolution, 21, 123–129.
Pereira, H. M., & Daily, G. C. (2006). Modeling biodiversity dynamics in countryside landscapes. Ecology, 87, 1877–1885.
Pereira, H. M., Leadley, P. W., Proenca, V., Alkemade, R., Scharlemann, J. P. W., Fernandez-Manjarres, J. F., et al. (2010). Scenarios for global biodiversity in the 21st century. Science, 330, 1496–1501.
Porzig, E. L., Seavy, N. E., Gardali, T., Geupel, G. R., Holyoak, M., & Eadie, J. M. (2014). Habitat suitability through time: Using time series and habitat models to understand changes in bird density. Ecosphere, 5, 16.
Prober, S. M., Hilbert, D. W., Ferrier, S., Dunlop, M., & Gobbett, D. (2012). Combining community-level spatial modelling and expert knowledge to inform climate adaptation in temperate grassy eucalypt woodlands and related grasslands. Biodiversity and Conservation, 21, 1627–1650.
Proenca, V., & Pereira, H. M. (2013). Species-area models to assess biodiversity change in multi-habitat landscapes: The importance of species habitat affinity. Basic and Applied Ecology, 14, 102–114.
Rapacciuolo, G., Roy, D. B., Gillings, S., & Purvis, A. (2014). Temporal validation plots: Quantifying how well correlative species distribution models predict species’ range changes over time. Methods in Ecology and Evolution, 5, 407–420.
Rios-Munoz, C. A., & Navarro-Siguenza, A. G. (2009). Effects of land use change on the hypothetical habitat availability for Mexican parrots. Ornitologia Neotropical, 20, 491–509.
Santika, T., McAlpine, C. A., Lunney, D., Wilson, K. A., & Rhodes, J. R. (2014). Modelling species distributional shifts across broad spatial extents by linking dynamic occupancy models with public-based surveys. Diversity and Distributions, 20, 786–796.
Sayre, R., Dangermond, J., Frye, C., Vaughan, R., Aniello, P., & Breyer, S. et al. (2014) A new map of global ecological land units—An ecophysiographic stratification approach. Washington, D.C.: Association of American Geographers. http://www.aag.org/galleries/default-file/AAG_Global_Ecosyst_bklt72.pdf
Scholes, R. J., & Biggs, R. (2005). A biodiversity intactness index. Nature, 434, 45–49.
Shmueli, G. (2010). To explain or to predict? Statistical Science, 25, 289–310.
Skidmore, A. K., Pettorelli, N., Coops, N. C., Geller, G. N., Hansen, M., Lucas, R., et al. (2015). Agree on biodiversity metrics to track from space. Nature, 523, 403–405.
Smith, A. M. S., Falkowski, M. J., Greenberg, J. A., & Tinkham, W. T. (2014). Remote sensing of vegetation structure, function, and condition: Special issue. Remote Sensing of Environment, 154, 319–321.
Soberon, J., & Peterson, A. T. (2009). Monitoring biodiversity loss with primary species-occurrence data: Toward national-level indicators for the 2010 target of the convention on biological diversity. AMBIO, 38, 29–34.
Souza, D. M., Teixeira, R. F. M., & Ostermann, O. P. (2015). Assessing biodiversity loss due to land use with life cycle assessment: Are we there yet? Global Change Biology, 21, 32–47.
Stoms, D. M., Davis, F. W., & Cogan, C. B. (1992). Sensitivity of wildlife habitat models to uncertainties in GIS data. Photogrammetric Engineering and Remote Sensing, 58, 843–850.
Tingley, M. W., & Beissinger, S. R. (2009). Detecting range shifts from historical species occurrences: new perspectives on old data. Trends in Ecology & Evolution, 24, 625–633.
Tuanmu, M. N., & Jetz, W. (2014). A global 1-km consensus land-cover product for biodiversity and ecosystem modelling. Global Ecology and Biogeography, 23, 1031–1045.
Turak, E., Ferrier, S., Barrett, T., Mesley, E., Drielsma, M., Manion, G., et al. (2011). Planning for the persistence of river biodiversity: Exploring alternative futures using process-based models. Freshwater Biology, 56, 39–56.
Turner, W. (2014). Sensing biodiversity. Science, 346, 301–302.
van Strien, A. J., van Swaay, C. A. M., & Termaat, T. (2013). Opportunistic citizen science data of animal species produce reliable estimates of distribution trends if analysed with occupancy models. Journal of Applied Ecology, 50, 1450–1458.
van Vuuren, D. P., Kok, M. T. J., Girod, B., Lucas, P. L., & de Vries, B. (2012). Scenarios in global environmental assessments: Key characteristics and lessons for future use. Global Environmental Change-Human and Policy Dimensions, 22, 884–895.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution-Noncommercial 2.5 License (http://creativecommons.org/licenses/by-nc/2.5/) which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
The images or other third party material in this chapter are included in the work’s Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work’s Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2017 The Author(s)
About this chapter
Cite this chapter
Ferrier, S., Jetz, W., Scharlemann, J. (2017). Biodiversity Modelling as Part of an Observation System. In: Walters, M., Scholes, R. (eds) The GEO Handbook on Biodiversity Observation Networks. Springer, Cham. https://doi.org/10.1007/978-3-319-27288-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-27288-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27286-3
Online ISBN: 978-3-319-27288-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)