Abstract
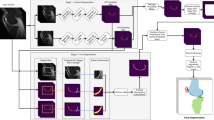
The morphological changes in knee cartilage (especially femoral and tibial cartilages) are closely related to the progression of knee osteoarthritis, which is expressed by magnetic resonance (MR) images and assessed on the cartilage segmentation results. Thus, it is necessary to propose an effective automatic cartilage segmentation model for longitudinal research on osteoarthritis. In this research, to relieve the problem of inaccurate discontinuous segmentation caused by the limited receptive field in convolutional neural networks, we proposed a novel position-prior clustering-based self-attention module (PCAM). In PCAM, long-range dependency between each class center and feature point is captured by self-attention allowing contextual information re-allocated to strengthen the relative features and ensure the continuity of segmentation result. The clutsering-based method is used to estimate class centers, which fosters intra-class consistency and further improves the accuracy of segmentation results. The position-prior excludes the false positives from side-output and makes center estimation more precise. Sufficient experiments are conducted on OAI-ZIB dataset. The experimental results show that the segmentation performance of combination of segmentation network and PCAM obtains an evident improvement compared to original model, which proves the potential application of PCAM in medical segmentation tasks. The source code is publicly available from link: https://github.com/PerceptionComputingLab/PCAMNet
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
References
Tack, A., Zachow, S.: Accurate automated volumetry of cartilage of knee using convolutional neural networks: data from the osteoarthritis initiative. In: 16th International Symposium on Biomedical Imaging, Venice, pp. 40–43. IEEE (2019)
Marinetti, A., et al.: Morphological MRI of knee cartilage: repeatability and reproducibility of damage evaluation and correlation with gross pathology examination. Eur. Radiol. 30(6), 3226–3235 (2019). https://doi.org/10.1007/s00330-019-06627-5
Gatti, A.A., Maly, M.R.: Automatic knee cartilage and bone segmentation using multi-stage convolutional neural networks: data from the osteoarthritis initiative. Magn. Reson. Mater. Phys., Biol. Med. 34(6), 859–875 (2021). https://doi.org/10.1007/s10334-021-00934-z
Ambellan, F., Tack., Ehlke, M., Zachow, S.: Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med. Image Anal. 52, 109–118 (2019)
Tan, C., Yan, Z., Zhang, S., Li, K., Metaxas, D.N.: Collaborative multi-agent learning for MR knee articular cartilage segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 282–290. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_32
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Milletari, F., Navab, N., Ahmadi, S.-A.: V-Net: fully convolutional neural networks for volumetirc medical image segmentation. In: 4th International Conference on 3D Vision, Stanford, pp. 565–571. IEEE (2016)
Sinha, A., Dolz, J.: Multi-scale self-guided attention for medical image segmentation. IEEE J. Biomed. Health Inform. 25(1), 121–130 (2021)
Zhao, H.-S., Shi, J.-P., Qi, X.-J., Wang, X.-G., Jia, J.-Y.: Pyramid scene parsing network. In: IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, pp. 6230–6239. IEEE (2017)
Zhang, F., et al.:ACFNet: attentional class feature network for semantic segmentation. In: International Conference on Computer Vision, Seoul, pp. 6797–6806 IEEE (2019)
Isensee, F., Jaeger, P.-F., Kohl, S.-A.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Method 18, 203–211 (2021)
Otter, N., Porter, M.A., Tillmann, U., Grindrod, P., Harrington, H.A.: A roadmap for the computation of persistent homology. EPJ Data Sci. 6(1), 1–38 (2017). https://doi.org/10.1140/epjds/s13688-017-0109-5
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant 62001144 and Grant 62001141, and by Science and Technology Innovation Committee of Shenzhen Municipality under Grant JCYJ20210324131800002 and RCBS20210609103820029.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Liang, D., Liu, J., Wang, K., Luo, G., Wang, W., Li, S. (2022). Position-Prior Clustering-Based Self-attention Module for Knee Cartilage Segmentation. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13435. Springer, Cham. https://doi.org/10.1007/978-3-031-16443-9_19
Download citation
DOI: https://doi.org/10.1007/978-3-031-16443-9_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16442-2
Online ISBN: 978-3-031-16443-9
eBook Packages: Computer ScienceComputer Science (R0)