Abstract
Objectives
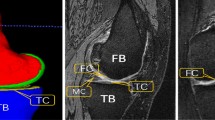
Accurate and efficient knee cartilage and bone segmentation are necessary for basic science, clinical trial, and clinical applications. This work tested a multi-stage convolutional neural network framework for MRI image segmentation.
Materials and methods
Stage 1 of the framework coarsely segments images outputting probabilities of each voxel belonging to the classes of interest: 4 cartilage tissues, 3 bones, 1 background. Stage 2 segments overlapping sub-volumes that include Stage 1 probability maps concatenated to raw image data. Using six fold cross-validation, this framework was tested on two datasets comprising 176 images [88 individuals in the Osteoarthritis Initiative (OAI)] and 60 images (15 healthy young men), respectively.
Results
On the OAI segmentation dataset, the framework produces cartilage segmentation accuracies (Dice similarity coefficient) of 0.907 (femoral), 0.876 (medial tibial), 0.913 (lateral tibial), and 0.840 (patellar). Healthy cartilage accuracies are excellent (femoral = 0.938, medial tibial = 0.911, lateral tibial = 0.930, patellar = 0.955). Average surface distances are less than in-plane resolution. Segmentations take 91 ± 11 s per knee.
Discussion
The framework learns to automatically segment knee cartilage tissues and bones from MR images acquired with two sequences, producing efficient, accurate quantifications at varying disease severities.






Similar content being viewed by others
References
Deshpande BR, Katz JN, Solomon DH, Yelin EH, Hunter DJ, Messier SP, Suter LG, Losina E (2016) Number of persons with symptomatic knee osteoarthritis in the US: impact of race and ethnicity, age, sex, and obesity: symptomatic knee OA in the US. Arthritis Care Res. https://doi.org/10.1002/acr.22897
Creamer P, Hochberg MC (1997) Osteoarthritis. Lancet 350:503–508
Kraus VB, Blanco FJ, Englund M, Karsdal MA, Lohmander LS (2015) Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthritis Cartilage 23:1233–1241
Hunter DJ, Altman RD, Cicuttini F, Crema MD, Duryea J, Eckstein F, Guermazi A, Kijowski R, Link TM, Martel-Pelletier J, Miller CG, Mosher TJ, Ochoa-Albíztegui RE, Pelletier J-P, Peterfy C, Raynauld J-P, Roemer FW, Totterman SM, Gold GE (2015) OARSI clinical trials recommendations: knee imaging in clinical trials in osteoarthritis. Osteoarthritis Cartilage 23:698–715
Conaghan PG, Hunter DJ, Maillefert JF, Reichmann WM, Losina E (2011) Summary and recommendations of the OARSI FDA osteoarthritis Assessment of Structural Change Working Group. Osteoarthritis Cartilage 19:606–610
Peterfy C, Woodworth T, Altman R (2006) Workshop for consensus on osteoarthritis imaging: MRI of the knee. Osteoarthritis Cartilage 14:44–45
Metcalfe AJ, Andersson ML, Goodfellow R, Thorstensson CA (2012) Is knee osteoarthritis a symmetrical disease? Analysis of a 12 year prospective cohort study. BMC Musculoskelet Disord. https://doi.org/10.1186/1471-2474-13-153
Pedoia V, Majumdar S, Link TM (2016) Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phys Biol Med 29:207–221
Duryea J, Neumann G, Brem MH, Koh W, Noorbakhsh F, Jackson RD, Yu J, Eaton CB, Lang P (2007) Novel fast semi-automated software to segment cartilage for knee MR acquisitions. Osteoarthritis Cartilage 15:487–492
Shim H, Chang S, Tao C, Wang J-H, Kwoh CK, Bae KT (2009) Knee cartilage: efficient and reproducible segmentation on high-spatial-resolution MR images with the semiautomated graph-cut algorithm method. Radiology 251:548–556
Peterfy CG, Schneider E, Nevitt M (2008) The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage 16:1433–1441
Shan L, Zach C, Charles C, Niethammer M (2014) Automatic atlas-based three-label cartilage segmentation from MR knee images. Med Image Anal 18:1233–1246
Ahn C, Bui TD, Lee Y, Shin J, Park H (2016) Fully automated, level set-based segmentation for knee MRIs using an adaptive force function and template: data from the osteoarthritis initiative. Biomed Eng Online. https://doi.org/10.1186/s12938-016-0225-7
Dodin P, Pelletier J, Martel-Pelletier J, Abram F (2010) Automatic human knee cartilage segmentation from 3-D magnetic resonance images. IEEE Trans Biomed Eng 57:2699–2711
Fripp J, Crozier S, Warfield SK, Ourselin S (2010) Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans Med Imaging 29:55–64
Dam EB, Lillholm M, Marques J, Nielsen M (2015) Automatic segmentation of high- and low-field knee MRIs using knee image quantification with data from the osteoarthritis initiative. J Med Imaging (Bellingham) 2:024001
Wang Q, Wu D, Lu L, Liu M, Boyer KL, Zhou SK (2014) Semantic Context Forests for Learning-Based Knee Cartilage Segmentation in 3D MR Images. In: Menze B, Langs G, Montillo A, Kelm M, Müller H, Tu Z (eds) Medical Computer Vision. Large Data in Medical Imaging. Springer International Publishing, Cham, pp 105–115
Prasoon A, Igel C, Loog M, Lauze F, Dam EB, Nielsen M (2013) Femoral cartilage segmentation in knee MRI scans using two stage voxel classification. In: Engineering in medicine and biology society (EMBC), 2013 35th annual international conference of the IEEE. IEEE, pp 5469–5472
Tamez-Pena JG, Farber J, Gonzalez PC, Schreyer E, Schneider E, Totterman S (2012) Unsupervised segmentation and quantification of anatomical knee features: data from the osteoarthritis initiative. IEEE Trans Biomed Eng 59:1177–1186
Yin Y, Zhang X, Williams R, Xiaodong Wu, Anderson DD, Sonka M (2010) LOGISMOS—layered optimal graph image segmentation of multiple objects and surfaces: cartilage segmentation in the knee joint. IEEE Trans Med Imaging 29:2023–2037
Shen D, Wu G, Suk H-I (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248
Ambellan F, Tack A, Ehlke M, Zachow S (2019) Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med Image Anal 52:109–118
Liu F (2018) SUSAN: segment unannotated image structure using adversarial network. Magn Reson Med. https://doi.org/10.1002/mrm.27627
Zhou Z, Zhao G, Kijowski R, Liu F (2018) Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 80:2759–2770
Norman B, Pedoia V, Majumdar S (2018) Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 288:177–185
Milletari F, Navab N, Ahmadi S-A (2016) V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV). IEEE, Stanford, CA, USA, pp 565–571 http://arxiv.org/abs/1606.04797v1
Yu L, Yang X, Chen H, Qin J, Heng P-A (2017) Volumetric ConvNets with Mixed Residual Connections for Automated Prostate Segmentation from 3D MR Images. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI’17). AAAI Press, pp 66–72
Zeng G, Zheng G (2019) 3D tiled convolution for effective segmentation of volumetric medical images. In: Shen D, Liu T, Peters TM, Staib LH, Essert C, Zhou S, Yap P-T, Khan A (eds) Medical image computing and computer assisted intervention—MICCAI 2019. Springer International Publishing, Cham, pp 146–154
Zhu Z, Xia Y, Xie L, Fishman EK, Yuille AL (2019) Multi-scale coarse-to-fine segmentation for screening pancreatic ductal adenocarcinoma. Prepring at arXiv:1807.02941 [cs]
Roth HR, Oda H, Zhou X, Shimizu N, Yang Y, Hayashi Y, Oda M, Fujiwara M, Misawa K, Mori K (2018) An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput Med Imaging Graph 66:90–99
Roth HR, Lu L, Lay N, Harrison AP, Farag A, Sohn A, Summers RM (2018) Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal 45:94–107
Pang S, Du A, He X, Díez J, Orgun MA (2019) Fast and accurate lung tumor spotting and segmentation for boundary delineation on CT slices in a coarse-to-fine framework. In: Gedeon T, Wong KW, Lee M (eds) Neural information processing. Springer International Publishing, Cham, pp 589–597
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Kayalibay B, Jensen G, van der Smagt P (2017) CNN-based segmentation of medical imaging data. Preprint at arXiv:1701.03056 [cs]
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In: 2015 IEEE international conference on computer vision (ICCV). IEEE, Santiago, Chile, pp 1026–1034
Kingma DP, Ba J (2017) Adam: a method for stochastic optimization. Preprint at arXiv:1412.6980 [cs]
Gatti AA (2018) NEURALSEG: state-of-the-art cartilage segmentation using deep learning–analyses of data from the osteoarthritis initiative. Abstracts from the 2018 OARSI World Congress on Osteoarthritis. Osteoarthritis and Cartilage, pp 47–48
Caliva F, Iriondo C, Martinez AM, Majumdar S, Pedoia V (2019) Distance map loss penalty term for semantic segmentation. Preprint at arXiv:1908.03679 [cs, eess]
Kellgren JH, Lawrence JS (1957) Radiological assessment of osteo-arthrosis. Ann Rheum Dis 16:494–502
Schneider E, NessAiver M, White D, Purdy D, Martin L, Fanella L, Davis D, Vignone M, Wu G, Gullapalli R (2008) The osteoarthritis initiative (OAI) magnetic resonance imaging quality assurance methods and results. Osteoarthritis Cartilage 16:994–1004
Williams TG, Holmes AP, Bowes M, Vincent G, Hutchinson CE, Waterton JC, Maciewicz RA, Taylor CJ (2010) Measurement and visualisation of focal cartilage thickness change by MRI in a study of knee osteoarthritis using a novel image analysis tool. Br J Radiol 83:940–948
Gatti AA, Noseworthy MD, Stratford PW, Brenneman EC, Totterman S, Tamez-Peña J, Maly MR (2017) Acute changes in knee cartilage transverse relaxation time after running and bicycling. J Biomech 53:171–177
Hastie T, Tibshirani R, Friedman JH (2009) The elements of statistical learning: data mining, inference, and prediction, 2nd edn. Springer, New York
Desai AD, Caliva F, Iriondo C, Mortazi A, Jambawalikar S, Bagci U, Perslev M, Igel C, Dam EB, Gaj S, Yang M, Li X, Deniz CM, Juras V, Regatte R, Gold GE, Hargreaves BA, Pedoia V, Chaudhari AS (2021) The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset. Radiology: Artificial Intelligence 3:e200078
Panfilov E, Tiulpin A, Klein S, Nieminen MT, Saarakkala S (2019) Improving robustness of deep learning based knee MRI segmentation: mixup and adversarial domain adaptation. In: 2019 IEEE/CVF international conference on computer vision workshop (ICCVW). IEEE, Seoul, Korea (South), pp 450–459
Gaj S, Yang M, Nakamura K, Li X (2020) Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks. Magn Reson Med 84:437–449
Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2017) Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging: Deep Learning Approach for Segmenting MR Image. Magn Reson Med. https://doi.org/10.1002/mrm.26841
Ioffe S (2017) Batch renormalization: towards reducing minibatch dependence in batch-normalized models. Preprint at arXiv:1702.03275 [cs]
Masters D, Luschi C (2018) Revisiting small batch training for deep neural networks. Preprint at arXiv:1804.07612 [cs, stat]
Lian X, Liu J (2019) Revisit batch normalization: new understanding and refinement via composition optimization. In: Proceedings of machine learning research. pp 3254–3263
Deng J, Dong W, Socher R, Li L-J, Kai Li, Li Fei-Fei (2009) ImageNet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE, Miami, FL, pp 248–255
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25:1097–1105
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical Image Analysis 3661-78. https://doi.org/10.1016/j.media.2016.10.004
Acknowledgements
We would like to acknowledge Google for providing cloud compute credits used to conduct the experiments. The Osteoarthritis Initiative (OAI) is a public-private partnership funded by the National Institutes of Health (NIH) and private partners including Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. This manuscript was prepared using an OAI public use data set and does not reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Funding
A. A. Gatti was supported by an Ontario Graduate Scholarship, The Arthritis Society, and a Mitacs Accelerate Entrepreneur award. M.R. Maly holds The Arthritis Society Stars Mid-Career Development Award funded by the Canadian Institutes of Health Research-Institute of Musculoskeletal Health and Arthritis and an NSERC Discovery grant that supported this work (MRM: 353715).
Author information
Authors and Affiliations
Contributions
AAG contributed to study conception and design, acquisition of data, analysis and interpretation of data, drafting of manuscript, and critical revision. MRM contributed to acquisition of data, interpretation of data, drafting of manuscript, and critical revision.
Corresponding author
Ethics declarations
Conflict of interest
A. A Gatti is the founder of NeuralSeg, Ltd. There are no other conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gatti, A.A., Maly, M.R. Automatic knee cartilage and bone segmentation using multi-stage convolutional neural networks: data from the osteoarthritis initiative. Magn Reson Mater Phy 34, 859–875 (2021). https://doi.org/10.1007/s10334-021-00934-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-021-00934-z