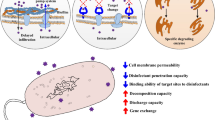
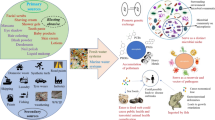
Abstract
Microplastic pollution is a big and rapidly growing environmental problem. Although the direct effects of microplastic pollution are increasingly studied, the indirect effects are hardly investigated, especially in the context of spreading of disease and antibiotic resistance genes, posing an apparent hazard for human health. Microplastic particles provide a hydrophobic surface that provides substrate for attachment of microorganisms and readily supports formation of microbial biofilms. Pathogenic bacteria such as fish pathogens Aeromonas spp., Vibrio spp., and opportunistic human pathogens like Escherichia coli are present in these biofilms. Moreover, some of these pathogens are shown to be multidrug resistant. The presence of microplastics is known to enhance horizontal gene transfer in bacteria and thus, may contribute to dissemination of antibiotic resistance. Microplastics can also adsorb toxic chemicals like antibiotics and heavy metals, which are known to select for antibiotic resistance. Microplastics may, thus, serve as vectors for transport of pathogens and antibiotic resistance genes in the aquatic environment. In this book chapter, we provide background information on microplastic biofouling (“plastisphere concept”), discuss the relationship between microplastic and antibiotic resistance, and identify knowledge gaps and directions for future research.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
9.1 Introduction
Microplastic (<5 mm, GESAMP 2019) pollution is a widespread and global environmental problem that is projected to increase in upcoming decades creating significant challenges for its management and prevention (Borrelle et al. 2020; Jambeck et al. 2015). Transport of microplastic from land via headwater streams and large rivers to the ocean (Hurley et al. 2018; Jambeck et al. 2015; van Wijnen et al. 2019) is an important component of the microplastic pollution cycle , and plastic particles can now be found globally throughout all ecosystem components including the atmosphere, terrestrial landscapes, aquatic freshwater and marine environments, and all types of biota including seafood species commonly consumed by humans (Bank and Hansson 2019).
Microplastics represent a novel substrate for marine bacteria including both fish and human pathogens (Dang and Lovell 2016; McCormick et al. 2014; Zettler et al. 2013) and are also a reservoir for metal resistance and antibiotic resistance genes. The role of microplastics in the spread of antibiotic resistance is a relatively new research topic that has garnered significant interest by scientists (Bank et al. 2020; Bowley et al. 2021; Guo et al. 2020; Parthasarathy et al. 2019; Radisic et al. 2020). The indirect effects of microplastics have not been well studied especially in the context of seafood safety and global food security, and these effects may pose a significant hazard for human health regarding the spread of disease (Bank et al. 2020; Guo et al. 2020). The specific objectives of this chapter were to (1) provide background information on microplastic biofouling and describe the concept of the “plastisphere” (Zettler et al. 2013), (2) discuss the relationship of microplastic and antibiotic resistance, and (3) identify knowledge gaps and directions for future research.
9.2 The Plastisphere Concept
One of the critical mechanisms of the microplastic antibiotic resistance connection is the “plastisphere” concept. This concept was originally presented in the seminal paper by Zettler et al. (2013) who reported that microbial communities attached to plastic debris were diverse and composed of heterotrophs, autotrophs, predators, and symbionts and were distinct from the surrounding marine waters. These plastic particle surfaces represented a novel substrate and/or ecological habitat within the water column and on the surface of the open ocean (Amaral-Zettler et al. 2015, 2020; Bowley et al. 2021; Oberbeckmann et al. 2018; Wright et al. 2020; Zettler et al. 2013). Microplastic particles have hydrophobic surfaces, with no net charge, upon entering the ocean as virgin artificial materials; however, they can quickly become colonized by microbial biofilms (Bowley et al. 2021; Wright et al. 2020; Zettler et al. 2013). The development of this concept was important for forming scientific questions regarding the overall direct and indirect impacts of microplastic pollution primarily because of the long residence time of microplastic in the environment and the potential for long-range transport and the associated risks of transfer of pathogens and disease (Bowley et al. 2021). Pathogenic microbes such as Vibrio spp. have been reported in high abundance within the plastisphere (Amaral-Zettler et al. 2020; Bowley et al. 2021; Zettler et al. 2013; Zhang et al. 2020) and although not all vibrios are pathogenic, they often prefer lower salinity found in coastal and estuarine areas, thus highlighting the importance of the plastisphere regarding its distribution, abundance, fate, and transport (Bowley et al. 2021; Thompson et al. 2004). These identified risks and the processes related to microplastic and microbe interactions are complex and are influenced by ocean currents (Hale et al. 2020), sources, fate and transport dynamics, trophic transfer and food web complexity, horizontal gene transfer and attachment properties (Arias-Andres et al. 2019), buoyancy and sinking properties of microplastics, variation, and uptake by farmed (Sun et al. 2020a) and wild seafood taxa, leading to subsequent human exposures (Bowley et al. 2021; Zhou et al. 2020).
9.3 Antibiotic Resistance
The introduction of antibiotics for the treatment of infectious disease is one of the most important advances in healthcare. The global spread of resistance in bacteria, particularly in human pathogens, presents major challenges for treatment and preventing the spread of infections (Ventola 2015). Annually, in the European Union/European Economic Area, an estimated more than 33,000 deaths and more than 800,000 cases of “impacted life-years” are attributable to infections caused by antibiotic-resistant pathogens, with direct and indirect estimated costs of more than 1.5 B€ (Cassini et al. 2019). The World Health Organization (WHO) has predicted the advent of infectious diseases for which no antibiotic treatment will be available (WHO 2019).
Antibiotic resistance is a natural phenomenon. Misuse and over use of antibiotics has led to the development, selection, and global spread of antibiotic resistance (Roberts and Zembower 2020). Selection pressure from the presence of antibiotics or other antimicrobial compounds like heavy metals and biocides leads to the enrichment of antibiotic-resistant bacteria and antibiotic resistance genes (ARGs) in bacteria from humans, animals, and the environment (Francino 2016; Gullberg et al. 2014; Marathe et al. 2013; Seiler and Berendonk 2012). Horizontal gene transfer (HGT) is a fundamental force driving bacterial evolution and contributes to the dissimilation of ARGs in both clinical and environmental bacteria (Boto 2010; Emamalipour et al. 2020; Jain et al. 2003). Antimicrobial compounds like antibiotics, biocides, and heavy metals can drive the development of antibiotic resistance and stimulate horizontal transfer of antibiotic resistance genes (Andersson and Hughes 2014; Jutkina et al. 2018; Zhang et al. 2018), thus aiding selection and dissemination of antibiotic resistance.
9.4 Microplastics and Antibiotic Resistance
Microorganisms attach themselves to surfaces forming a complex matrix of biopolymers and microbial cells known as biofilm (Dang and Lovell 2016). Formation of biofilms protect bacteria from unfavorable conditions in the environment (Donlan 2002). Biofilms are ubiquitous in aquatic environments and play an important role in various biological and ecological processes (Guo et al. 2018). Aquatic biofilms serve as a sink for various contaminants, like heavy metals, and antibiotics that are known to select for antibiotic resistance and stimulate horizontal transfer of antibiotic resistance genes (Gullberg et al. 2014; Guo et al. 2018; Jutkina et al. 2018; Richard et al. 2019). Accordingly, antibiotic resistance genes have been detected in natural aquatic biofilms (Balcázar et al. 2015; Guo et al. 2018).
Microplastic particles provide a hydrophobic surface that readily supports formation of microbial biofilms, where environmental conditions are the main drivers of biofilm formation (Oberbeckmann et al. 2018; Rummel et al. 2017). Pathogenic bacteria such as fish pathogens Aeromonas spp., Vibrio spp., and opportunistic human pathogens like E. coli can invariably be present in these biofilms (Kirstein et al. 2016; Silva et al. 2019; Viršek et al. 2017). Microplastics can selectively enrich both antibiotics and antibiotic-resistant bacteria on their surfaces in landfill leachates, freshwater, as well as in sea water (Su et al. 2020; Sun et al. 2020b; Wang et al. 2020; Wu et al. 2019). Thus, microplastics may serve as a vector for transport of pathogens in the aquatic environment.
Several methods have been used for detecting and quantifying ARGs associated with marine plastics including selective isolation of resistant bacteria and phenotypic antibiotic sensitivity testing, whole genome sequencing, shotgun metagenomics, and quantitative polymerase chain reaction (qPCR). Culture-based methods involving isolation of bacteria on a culture media followed by antibiotic sensitivity testing is a traditional approach used for studying antibiotic resistance (Khan et al. 2019). Zhang et al. (2020) carried out isolation and characterization of antibiotic-resistant marine bacteria from microplastic particles collected from marine aquaculture sites using a combination of seven antibiotics and a non-selective media. They showed presence of several multidrug-resistant marine bacteria including pathogenic Vibrio species on microplastics (Zhang et al. 2020). In contrast, other studies carried out selective isolation of pathogens like Vibrio spp. (Laverty et al. 2020) and E. coli (Song et al. 2020) showing presence of multidrug-resistant pathogens on marine microplastics. Recently, a study reported whole genome sequences (WGS) of antibiotic-resistant fish pathogens isolated from marine plastics (Radisic et al. 2020). With the advent of next-generation sequencing technology, WGS analysis of pathogens has become common and affordable tool for genotyping and epidemiology in clinics (Quainoo et al. 2017). WGS analyses are effective in elucidating the total metabolic potential of microorganisms and understanding the genetic basis of antibiotic resistance (Grevskott et al. 2020; Hendriksen et al. 2019). Although this is true, WGS data on microplastic-associated bacteria is largely lacking.
Only a small proportion of bacteria present in an environment can be cultivated in the lab. This limits detection and quantification of antibiotic resistance genes present in uncultivable bacteria using traditional methods (Lloyd et al. 2018; Stewart 2012). Methods like qPCR analysis or shotgun metagenomics, that use the total genomic DNA extracted from a given sample, partly overcome this limitation. Using qPCR, Wang et al. (2020) showed enrichment of ARGs like sul1, tetA, tetC, tetX, and ermE on plastic particles in both freshwater and sea water (Wang et al. 2020), while another study showed selective enrichment of strB, blaTEM, ermB, tetM, and tetQ on microplastic particles in landfill leachates (Shi et al. 2020). These studies selected a limited number of ARGs for their analysis. In contrast, using recently developed high-throughput qPCR screening that can analyze more than 200 ARGs, Lu et al. (2020) showed presence of between 34 and 43 different ARGs on the surface of microplastic particles collected from vegetable soil (Lu et al. 2020).
Shotgun metagenomics gives an overview of the total bacteria and associated genes present in a given sample (Simon and Daniel 2011). Using this method Yang et al. (2019) found a total of 64 ARG subtypes that provide resistance against 13 different classes of antibiotics on macroplastics and microplastics collected from the North Pacific Gyre. Along with enrichment of ARGs, the study also found enrichment of metal resistance genes on microplastics (Yang et al. 2019). This study and several of the earlier described studies show presence of clinically important ARGs, like sul1, tetA, tetC, tetX, ermE, aac(3), macB, and blaTEM, that are invariably found in human pathogens, on microplastic particles (Alcock et al. 2020), thus suggesting that microplastics in the environment act as reservoirs for clinically important antibiotic resistance genes.
Microplastics originate from a variety of processes and invariably ends up in the marine environment via streams and large rivers (Hurley et al. 2018; Jambeck et al. 2015). High levels of microplastics reach the wastewater treatment plants (WWTP) (Dris et al. 2015). Although most of the microplastics are removed during primary and secondary waste treatment, smaller microplastics may still be present in the treated effluents (Talvitie et al. 2017). Treated effluents have low concentrations of microplastic particles but the high volume of effluents released may leads to considerable contamination of the aquatic ecosystem (Murphy et al. 2016; Talvitie et al. 2017). WWTPs receive municipal and/or hospital waste which invariably contains both human pathogens and clinically important antibiotic resistance genes (Le et al. 2016; Marathe et al. 2017, 2018, 2019; Rizzo et al. 2013). Treated effluents from waste water treatment plants are recognized as one of the major sources of environmental pollution with antibiotic resistance genes and resistant pathogens (Alexander et al. 2020; Czekalski et al. 2014; Karkman et al. 2019). The presence of microplastic particles in waste water effluents, thus, presents opportunities for antibiotic-resistant pathogens to colonize and form biofilms on plastic particles. This may lead to further dissemination of resistance in the marine environment via microplastics. Although this is true, there is limited knowledge on the impact of microplastics from treated effluents from WWTP on dissemination of ARGs in the aquatic environment.
Microplastic particles adsorb several chemicals like antibiotics, biocides, and heavy metals (Chen et al. 2020; Godoy et al. 2019; Mammo et al. 2020; Wang et al. 2020). The presence of antibiotics and active metabolites from such agents in the environment leads to selection of multidrug resistance among both clinical and environmental bacteria. Similarly, biocides and heavy metals like copper and mercury are known to co-select for antibiotic resistance (Francino 2016; Gullberg et al. 2011, 2014; Imran et al. 2019; Marathe et al. 2013; Seiler and Berendonk 2012). Adsorption of these chemicals on plastic surfaces containing microbial biofilm may lead to selection pressure in the plastisphere, resulting in active selection of antibiotic resistance on microplastic surfaces. In accordance, Imran et al. (2019) has concluded that co-contamination with microplastics and heavy metals results in development and spread of multiple drug-resistant human pathogens through co-selection mechanisms (Imran et al. 2019). Studies have shown that very low levels of antibiotics and biocides not only can select for antibiotic resistance but also can induce horizontal transfer of ARGs (Gullberg et al. 2011; Jutkina et al. 2018; Zhang et al. 2018). Moreover, bacteria in biofilms are more efficient in horizontal gene transfer compared to planktonic bacteria (Abe et al. 2020). Accordingly, studies have shown increased horizontal gene transfer in presence of microplastics via conjugation (Arias-Andres et al. 2018, 2019). Although extensive research on selection of resistance and promotion of horizontal gene transfer by antimicrobial compounds has been carried out, there is limited knowledge on the effect of adsorbed chemicals on plastisphere bacteria, especially, with reference to selection and transfer of antibiotic resistance genes on microplastic particles.
9.5 Conclusions and Directions for Future Research
Microplastics are emerging pollutants that have been detected in a range of environments. With the current trend of plastic consumption and its global production, the environmental pollution and related environmental effects caused by microplastics are expected to increase (Borrelle et al. 2020). Microplastics provide surfaces for the microorganisms to form biofilms (plastisphere) (Zettler et al. 2013). The processes and mechanisms involved in biofilm formation on microplastics are largely unclear. In-depth studies on deciphering the succession of microbes and understanding the effect of different factors that may influence biofilm formation on microplastic particles, such as the environmental conditions and the age of microplastic particles are needed (Su et al. 2020; Yang et al. 2020). Moreover, there are a limited number of studies reporting WGS of bacteria associated with microplastics (Li et al. 2019; Radisic et al. 2021). Bacteria associated with microplastics may play different ecological roles and could also be useful for bioremediation (Debroas et al. 2017). Hence, understanding the metabolic potential of bacteria in plastisphere using WGS is necessary.
Studies have investigated the composition of biofilms on microplastics and shown presence of both fish and human pathogens as well as clinically important antibiotic resistance genes (Dong et al. 2021). However, the risks associated with presence of pathogens in terms of human or fish exposure and the ability of microplastic-associated pathogens for causing infections is not fully understood. In-depth risk assessment studies on the effect of pathogen carrying microplastics on fish and human health are thus warranted. Microplastics originating in different compartments like WWTPs or aquaculture sites may carry different microbiota. WWTPs and aquaculture sites usually have presence of both antibiotic-resistant pathogens and microplastics (Cabello et al. 2016; Rodriguez-Mozaz et al. 2015). There is invariably selection pressure due to presence of antibiotics or biocides along with presence of resistant bacteria in these sites (Cabello et al. 2016; Edo et al. 2020; Yang et al. 2014). These environments could play an important role in enrichment and dispersal of pathogenic bacteria and ARGs to the marine ecosystem via microplastics. Although microplastics have been shown to increase HGT (Arias-Andres et al. 2018, 2019), the impact of microplastics on evolution and dissemination of antibiotic resistance in pathogens and environmental bacteria is largely unknown. In order to understand the indirect effects of microplastics, the relationship and interactions between microplastics and ARGs, as well as the impact of their association on aquatic environment especially on marine environment and sea food safety, needs to be further assessed. Holistic multidisciplinary studies on fate, migration, and potential environmental risks posed by microplastics through dissemination and evolution of antibiotic resistance are needed in the future, for better understanding the indirect effects of microplastic pollution.
References
Abe K, Nomura N, Suzuki S (2020) Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol 96(5):fiaa031
Alcock BP, Raphenya AR, Lau TT, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48(D1):D517–D525
Alexander J, Hembach N, Schwartz T (2020) Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci Rep 10(1):1–9
Amaral-Zettler LA, Zettler ER, Mincer TJ (2020) Ecology of the plastisphere. Nat Rev Microbiol 18(3):1–13
Amaral-Zettler LA, Zettler ER, Slikas B, Boyd GD, Melvin DW, Morrall CE, Proskurowski G, Mincer TJ (2015) The biogeography of the Plastisphere: implications for policy. Front Ecol Environ 13(10):541–546
Andersson DI, Hughes D (2014) Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12(7):465–478
Arias-Andres M, Klümper U, Rojas-Jimenez K, Grossart H-P (2018) Microplastic pollution increases gene exchange in aquatic ecosystems. Environ Pollut 237:253–261
Arias-Andres M, Rojas-Jimenez K, Grossart H-P (2019) Collateral effects of microplastic pollution on aquatic microorganisms: an ecological perspective. TrAC Trends Anal Chem 112:234–240
Balcázar JL, Subirats J, Borrego CM (2015) The role of biofilms as environmental reservoirs of antibiotic resistance. Front Microbiol 6:1216
Bank MS, Hansson SV (2019) The plastic cycle: a novel and holistic paradigm for the Anthropocene. Environ Sci Technol 53(13):7177–7179
Bank MS, Ok YS, Swarzenski PW (2020) Microplastic’s role in antibiotic resistance. Science 369(6509):1315
Borrelle SB, Ringma J, Law KL, Monnahan CC, Lebreton L, McGivern A, Murphy E, Jambeck J, Leonard GH, Hilleary MA (2020) Predicted growth in plastic waste exceeds efforts to mitigate plastic pollution. Science 369(6510):1515–1518
Boto L (2010) Horizontal gene transfer in evolution: facts and challenges. Proc Biol Sci 277(1683):819–827
Bowley J, Baker-Austin C, Porter A, Hartnell R, Lewis C (2021) Oceanic hitchhikers–assessing pathogen risks from marine microplastic. Trends Microbiol 29(2):107–116
Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ (2016) Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis 16(7):e127–e133
Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M (2019) Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19(1):56–66
Chen X, Gu X, Bao L, Ma S, Mu Y (2020) Comparison of adsorption and desorption of triclosan between microplastics and soil particles. Chemosphere 263:127947
Czekalski N, Díez EG, Bürgmann H (2014) Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J 8(7):1381–1390
Dang H, Lovell CR (2016) Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80(1):91–138
Debroas D, Mone A, Ter Halle A (2017) Plastics in the North Atlantic garbage patch: a boat-microbe for hitchhikers and plastic degraders. Sci Total Environ 599:1222–1232
Dong H, Chen Y, Wang J, Zhang Y, Zhang P, Li X, Zou J, Zhou A (2021) Interactions of microplastics and antibiotic resistance genes and their effects on the aquaculture environments. J Hazard Mater 403:123961
Donlan RM (2002) Biofilms: microbial life on surfaces. Emerg Infect Dis 8(9):881
Dris R, Gasperi J, Rocher V, Saad M, Renault N, Tassin B (2015) Microplastic contamination in an urban area: a case study in greater Paris. Environ Chem 12(5):592–599
Edo C, González-Pleiter M, Leganés F, Fernández-Piñas F, Rosal R (2020) Fate of microplastics in wastewater treatment plants and their environmental dispersion with effluent and sludge. Environ Pollut 259:113837
Emamalipour M, Seidi K, Vahed SZ, Jahanban-Esfahlan A, Jaymand M, Majdi H, Amoozgar Z, Chitkushev L, Javaheri T, Jahanban-Esfahlan R (2020) Horizontal gene transfer: from evolutionary flexibility to disease progression. Front Cell Dev Biol 8:229
Francino M (2016) Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 6:1543
Godoy V, Blázquez G, Calero M, Quesada L, Martín-Lara M (2019) The potential of microplastics as carriers of metals. Environ Pollut 255:113363
GESAMP (2019) Guidelines or the monitoring and assessment of plastic litter and microplastics in the ocean (Kershaw P.J., Turra A. and Galgani F. eds), IMO/FAO/UNESCO-IOC/UNIDO/WMO/IAEA/UN/UNEP/UNDP/ISA Joint Group of Experts on the Scientific Aspects of Marine Environmental Protection). Rep. Stud. GESAMP No. 99, 130p
Grevskott DH, Salvà-Serra F, Moore ERB, Marathe NP (2020) Nanopore sequencing reveals genomic map of CTX-M-type extended spectrum β-lactamases carried by Escherichia coli strains isolated from blue mussels (Mytilus edulis) in Norway. BMC Microbiol 20:134
Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI (2014) Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 5(5):e01918-01914–e01918-01914
Gullberg E, Cao S, Berg OG, Ilback C, Sandegren L, Hughes D, Andersson DI (2011) Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7(7):e1002158
Guo X-P, Sun X-L, Chen Y-R, Hou L, Liu M, Yang Y (2020) Antibiotic resistance genes in biofilms on plastic wastes in an estuarine environment. Sci Total Environ 745:140916
Guo X-P, Yang Y, Lu D-P, Niu Z-S, Feng J-N, Chen Y-R, Tou F-Y, Garner E, Xu J, Liu M (2018) Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res 129:277–286
Hale RC, Seeley ME, La Guardia MJ, Mai L, Zeng EY (2020) A global perspective on microplastics. J Geophys Res Oceans 125(1):e2018JC014719
Hendriksen RS, Bortolaia V, Tate H, Tyson G, Aarestrup FM, McDermott P (2019) Using genomics to track global antimicrobial resistance. Front Public Health 7:242
Hurley R, Woodward J, Rothwell JJ (2018) Microplastic contamination of river beds significantly reduced by catchment-wide flooding. Nat Geosci 11(4):251–257
Imran M, Das KR, Naik MM (2019) Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere 215:846–857
Jain R, Rivera MC, Moore JE, Lake JA (2003) Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol 20(10):1598–1602
Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, Narayan R, Law KL (2015) Plastic waste inputs from land into the ocean. Science 347(6223):768–771
Jutkina J, Marathe N, Flach C-F, Larsson D (2018) Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci Total Environ 616:172–178
Karkman A, Pärnänen K, Larsson DJ (2019) Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat Commun 10(1):1–8
Khan ZA, Siddiqui MF, Park S (2019) Current and emerging methods of antibiotic susceptibility testing. Diagnostics 9(2):49
Kirstein IV, Kirmizi S, Wichels A, Garin-Fernandez A, Erler R, Löder M, Gerdts G (2016) Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp on microplastic particles. Mar Environ Res 120:1–8
Laverty AL, Primpke S, Lorenz C, Gerdts G, Dobbs FC (2020) Bacterial biofilms colonizing plastics in estuarine waters, with an emphasis on Vibrio spp. and their antibacterial resistance. PLoS One 15(8):e0237704
Le T-H, Ng C, Chen H, Yi XZ, Koh TH, Barkham TMS, Zhou Z, Gin KY-H (2016) Occurrences and characterization of antibiotic resistant bacteria and genetic determinants of hospital wastewaters in a tropical country. Antimicrob Agents Chemother 60(12):01556–01516
Li Q, Xu X, He C, Zheng L, Gao W, Sun C, Li J, Gao F (2019) Complete genome sequence of a quorum-sensing bacterium, Oceanicola sp. strain D3, isolated from a microplastic surface in coastal water of Qingdao. China Microbiol Resour Announc 8(40):e01022–e01019
Lloyd KG, Steen AD, Ladau J, Yin J, Crosby L (2018) Phylogenetically novel uncultured microbial cells dominate earth microbiomes. MSystems 3(5):e00055–e00018
Lu X-M, Lu P-Z, Liu X-P (2020) Fate and abundance of antibiotic resistance genes on microplastics in facility vegetable soil. Sci Total Environ 709:136276
Mammo F, Amoah I, Gani K, Pillay L, Ratha S, Bux F, Kumari S (2020) Microplastics in the environment: interactions with microbes and chemical contaminants. Sci Total Environ 743:140518
Marathe NP, Berglund F, Razavi M, Pal C, Dröge J, Samant S, Kristiansson E, Larsson DGJ (2019) Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 7(1):97
Marathe NP, Janzon A, Kotsakis SD, Flach C-F, Razavi M, Berglund F, Kristiansson E, Larsson DJ (2018) Functional metagenomics reveals a novel carbapenem-hydrolyzing mobile beta-lactamase from Indian river sediments contaminated with antibiotic production waste. Environ Int 112:279–286
Marathe NP, Pal C, Gaikwad SS, Jonsson V, Kristiansson E, Larsson DGJ (2017) Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. Water Res 1247:388–397
Marathe NP, Regina VR, Walujkar SA, Charan SS, Moore ERB, Larsson DGJ, Shouche YS (2013) A treatment plant receiving waste water from multiple bulk drug manufacturers is a reservoir for highly multi-drug resistant integron-bearing bacteria. PLoS One 8(10):e77310
McCormick A, Hoellein TJ, Mason SA, Schluep J, Kelly JJ (2014) Microplastic is an abundant and distinct microbial habitat in an urban river. Environ Sci Technol 48(20):11863–11871
Murphy F, Ewins C, Carbonnier F, Quinn B (2016) Wastewater treatment works (WwTW) as a source of microplastics in the aquatic environment. Environ Sci Technol 50(11):5800–5808
Oberbeckmann S, Kreikemeyer B, Labrenz M (2018) Environmental factors support the formation of specific bacterial assemblages on microplastics. Front Microbiol 8:2709
Parthasarathy A, Tyler AC, Hoffman MJ, Savka MA, Hudson AO (2019) Is plastic pollution in aquatic and terrestrial environments a driver for the transmission of pathogens and the evolution of antibiotic resistance? Environ Sci Technol 53(4):2
Quainoo S, Coolen JP, van Hijum SA, Huynen MA, Melchers WJ, van Schaik W, Wertheim HF (2017) Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev 30(4):1015–1063
Radisic V, Lunestad BT, Sanden M, Bank MS, Marathe NP (2021) Draft genome sequence of multidrug-resistant Pseudomonas protegens strain 11HC2, isolated from marine plastic collected from the west coast of Norway. Microbiol Resour Announc 10(2):e01285–e01220
Radisic V, Nimje PS, Bienfait AM, Marathe NP (2020) Marine plastics from Norwegian west coast carry potentially virulent fish pathogens and opportunistic human pathogens harboring new variants of antibiotic resistance genes. Microorganisms 8(8):1200
Richard H, Carpenter EJ, Komada T, Palmer PT, Rochman CM (2019) Biofilm facilitates metal accumulation onto microplastics in estuarine waters. Sci Total Environ 683:600–608
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D (2013) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360
Roberts SC, Zembower TR (2020) Global increases in antibiotic consumption: a concerning trend for WHO targets. Lancet Infect Dis 21(1):2
Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sànchez-Melsió A, Borrego CM, Barceló D, Balcázar JL (2015) Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res 69:234–242
Rummel CD, Jahnke A, Gorokhova E, Kühnel D, Schmitt-Jansen M (2017) Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ Sci Technol Lett 4(7):258–267
Seiler C, Berendonk TU (2012) Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 3:399
Shi J, Wu D, Su Y, Xie B (2020) Selective enrichment of antibiotic resistance genes and pathogens on polystyrene microplastics in landfill leachate. Sci Total Environ 142775
Silva MM, Maldonado GC, Castro RO, de Sá Felizardo J, Cardoso RP, dos Anjos RM, de Araújo FV (2019) Dispersal of potentially pathogenic bacteria by plastic debris in Guanabara Bay, RJ, Brazil. Mar Pollut Bull 141:561–568
Simon C, Daniel R (2011) Metagenomic analyses: past and future trends. Appl Environ Microbiol 77(4):1153–1161
Song J, Jongmans-Hochschulz E, Mauder N, Imirzalioglu C, Wichels A, Gerdts G (2020) The Travelling Particles: investigating microplastics as possible transport vectors for multidrug resistant E. coli in the Weser estuary (Germany). Sci Total Environ 720:137603
Stewart EJ (2012) Growing unculturable bacteria. J Bacteriol 194(16):4151–4160
Su Y, Zhang Z, Zhu J, Shi J, Wei H, Xie B, Shi H (2020) Microplastics act as vectors for antibiotic resistance genes in landfill leachate: the enhanced roles of the long-term aging process. Environ Pollut 270:116278
Sun X, Chen B, Xia B, Li Q, Zhu L, Zhao X, Gao Y, Qu K (2020a) Impact of mariculture-derived microplastics on bacterial biofilm formation and their potential threat to mariculture: a case in situ study on the Sungo Bay, China. Environ Pollut 262:114336
Sun Y, Cao N, Duan C, Wang Q, Ding C, Wang J (2020b) Selection of antibiotic resistance genes on biodegradable and non-biodegradable microplastics. J Hazard Mater 409:124979
Talvitie J, Mikola A, Setälä O, Heinonen M, Koistinen A (2017) How well is microlitter purified from wastewater?–a detailed study on the stepwise removal of microlitter in a tertiary level wastewater treatment plant. Water Res 109:164–172
Thompson FL, Iida T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol Rev 68(3):403–431
van Wijnen J, Ragas AM, Kroeze C (2019) Modelling global river export of microplastics to the marine environment: sources and future trends. Sci Total Environ 673:392–401
Ventola CL (2015) The antibiotic resistance crisis: part 1: causes and threats. Pharm Therapeutics 40(4):277
Viršek MK, Lovšin MN, Koren Š, Kržan A, Peterlin M (2017) Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida. Mar Pollut Bull 125(1–2):301–309
Wang S, Xue N, Li W, Zhang D, Pan X, Luo Y (2020) Selectively enrichment of antibiotics and ARGs by microplastics in river, estuary and marine waters. Sci Total Environ 708:134594
WHO (2019) No time to wait: securing the future from drug-resistant infections. World Health Organization, Geneva
Wright RJ, Erni-Cassola G, Zadjelovic V, Latva M, Christie-Oleza JA (2020) Marine plastic debris: a new surface for microbial colonization. Environ Sci Technol 54(19):11657–11672
Wu X, Pan J, Li M, Li Y, Bartlam M, Wang Y (2019) Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res 165:114979
Yang K, Chen Q-L, Chen M-L, Li H-Z, Liao H, Pu Q, Zhu Y-G, Cui L (2020) Temporal dynamics of antibiotic resistome in the plastisphere during microbial colonization. Environ Sci Technol 54(18):11322–11332
Yang Y, Li B, Zou S, Fang HH, Zhang T (2014) Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res 62:97–106
Yang Y, Liu G, Song W, Ye C, Lin H, Li Z, Liu W (2019) Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ Int 123:79–86
Zettler ER, Mincer TJ, Amaral-Zettler LA (2013) Life in the “plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol 47(13):7137–7146
Zhang Y, Gu AZ, Cen T, Li X, He M, Li D, Chen J (2018) Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ Pollut 237:74–82
Zhang Y, Lu J, Wu J, Wang J, Luo Y (2020) Potential risks of microplastics combined with superbugs: enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol Environ Saf 187:109852
Zhou W, Han Y, Tang Y, Shi W, Du X, Sun S, Liu G (2020) Microplastics aggravate the bioaccumulation of two waterborne veterinary antibiotics in an edible bivalve species: potential mechanisms and implications for human health. Environ Sci Technol 54(13):8115–8122
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this chapter
Cite this chapter
Marathe, N.P., Bank, M.S. (2022). The Microplastic-Antibiotic Resistance Connection. In: Bank, M.S. (eds) Microplastic in the Environment: Pattern and Process. Environmental Contamination Remediation and Management. Springer, Cham. https://doi.org/10.1007/978-3-030-78627-4_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-78627-4_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-78626-7
Online ISBN: 978-3-030-78627-4
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)