Abstract
As a “hallmark of cancer”, tumor-induced angiogenesis is one of the most important mechanisms of a tumor’s adaptation to changes in nutrient requirement. The angiogenic activity of certain tumors has been found to be predictive of a patient’s ultimate response to therapeutic intervention. This then begs the question if there are differences in vessel arrangement and corresponding convolutedness, between tumors that appear phenotypically similar, but respond differently to treatment. Even though textural radiomics and deep learning-based approaches have been shown to distinguish disease aggressiveness and assess therapeutic response, these descriptors do not specifically interpret differences in vessel characteristics. Moreover, most existing approaches have attempted to model disease characteristics just within tumor confines, or right outside, but do not consider explicit parenchymal vessel morphology. In this work, we introduce VaNgOGH (Vascular Network Organization via Hough transform), a new descriptor of architectural disorder of the tumor’s vascular network. We demonstrate the efficacy of VaNgOGH in two clinically challenging problems: (a) Predicting pathologically complete response (pCR) in breast cancer prior to treatment (BCa, N = 76) and (b) distinguishing benign nodules from malignant non-small cell lung cancer (LCa, N = 81). For both tasks, VaNgOGH had test area under the receiver operating characteristic curve (\(AUC_{BCa}\) = 0.75, \(AUC_{LCa}\) = 0.68) higher than, or comparable to, state of the art radiomic approaches (\(AUC_{BCa}\) = 0.75, \(AUC_{LCa}\) = 0.62) and convolutional neural networks (\(AUC_{BCa}\) = 0.67, \(AUC_{LCa}\) = 0.66). Interestingly, when a known radiomic signature was used in conjunction with VaNgOGH, \(AUC_{BCa}\) increased to 0.79.
N. Braman and P. Prasanna—Equal contribution. Research was supported by 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R21CA179327-01, R21CA195152-01, R01DK098503-02, 1C06-RR12463-01, PC120857, LC130463, T32EB007509, the DOD Prostate Cancer Idea Development Award, W81XWH-16-1-0329, the CCCC Pilot Grant, CCF VelaSano Grant, I-Corps program, OTF Program, and the Wallace H. Coulter Foundation Program in BME, CWRU. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Organized Vascular Network
- Hough Transform
- Radiomics Signature
- Convolutional Neural Network (CNN)
- Architectural Disorder
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
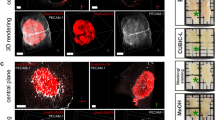
Angiogenesis, the process by which a tumor hijacks the body’s machinery for creating new vasculature in order to redirect blood flow to itself [1], plays an important role in determining tumor response to chemo- and radiotherapy. Stimulatory signals, such as vascular endothelial growth factor (VEGF) expression, result in neovascularization [2], ultimately leading to sprouting and irregular branching of blood vessels, or erratic angiogenesis. The associated tortuosity and leakiness directly affects the course of disease progression, and possibly its response to therapeutic interventions. For example, it has been qualitatively shown that temporal changes in vessel tortuosity on brain magnetic resonance imaging (MRI) are indicative of a favorable response to therapy [3]. A more convoluted tumor vasculature might constrict the delivery of therapeutic drugs to the lesion, thereby resulting in potentially worse prognosis and treatment response.
These qualitative observations therefore beg the question as to whether computerized analysis of tumor vasculature could (a) reveal differences in malignant and benign tumors as well as patients who undergo differential treatment response (see Fig. 1) and (b) whether these measurements could be translated into new imaging biomarkers of tumor diagnosis and treatment response.
In this work, we present a new image-based descriptor, Vascular Network Organization via Hough transform (VaNgOGH), which attempts to model the architectural disorder of a tumor’s vascular network by computing local measures of vessel-curvature in the Hough parameter space. VaNgOGH first looks at projections of vascular segmentations along different planes, in cartesian, as well as spherical coordinates. This is followed by localized Hough transforms to identify dominant peaks in the accumulator space. The applicability is demonstrated in two clinically challenging tasks: (a) predicting pathologic complete response (pCR) from non-response (non-pCR) to chemotherapy in breast cancer from pre-treatment dynamic contrast-enhance (DCE) MRI and (b) distinguishing benign granulomas from malignant adenocarcinomas from non-contrast computed tomography (CT) scans.
2 Previous Work and Novel Contributions
While there has been substantial interest in both radiomics and deep learning approaches for disease diagnosis as well as treatment response of tumors, most of these feature analysis approaches have been limited either to the tumor [4, 5] or the associated parenchyma [6, 7]. Recent work such as [6], which leveraged textural features of the tumor and the surrounding peri-tumoral tissue to predict therapeutic response from breast MRI, suggests the discriminating nature of the tumor microenvironment on imaging and the potential of extra-tumoral quantitative analysis. However, there is a lack of radiomic features capable of directly targeting biological aspects of the microenvironment, such as vascularity. VaNgOGH represents a novel approach for the characterization of chaotic vasculature associated with tumor-induced angiogenesis. In capturing morphology of the tumor-associated vascular network, VaNgOGH implicitly captures functional attributes of the tumor.
A unique advantage of VaNgOGH over other state-of-the-art quantitative metrics of vessel architecture is its capability to define abnormal vessel arrangement across multiple planes and projections, and relative to the tumor core and boundary itself. Specifically, VaNgOGH invokes Hough transformation to characterize the vessel network across multiple spatial representations, operating both in the cartesian domain, to capture disorder in the plane of image acquisition, and in the spherical domain, to capture deflections of neighboring vasculature towards the tumor centroid due to angiogenesis. In the latter operation, we further leverage the spherical coordinate space by computing VaNgOGH beyond the tumor within annular bands of increasing radius and summarize across regions in order to capture the magnitude of the tumor’s angiogenic influence. VaNgOGH features consist of the first order statistics of maximum Hough peak orientations computed in a sliding fashion across vessel projections summarizing vasculature orientation in the following domains: the XY-plane, distance from the tumor vs. azimuthal rotation, distance from the tumor vs. elevation angle, and azimuthal rotation vs. elevation angle.
In this work, we evaluated the approach on N = 157 breast and lung cancer patients in predicting treatment response and cancer presence, also comparing the approach against a convolutional neural network (CNN) and state-of-the-art textural radiomic measures of tumoral and peri-tumoral regions.
3 Methodology
3.1 Notation
We define an image scene \(\mathcal {I}\) as \(\mathcal {I}\,=\,(C,f)\), where \(\mathcal {I}\) is a spatial grid C of voxels \(c \in C\), in a 3-dimensional space, \(\mathbb {R}^{3}\). Each voxel, \(c \in C\) is associated with an intensity value f(c). \(\mathcal {I}_T\), and \(\mathcal {I}_P\) correspond to the intra-tumoral and surrounding peritumoral parenchyma sub-volumes within every \(\mathcal {I}\) respectively, such that \([\mathcal {I}_{T}, \mathcal {I}_{P}] \subset \mathcal {I}\). We further divide the sub-volume \(\mathcal {I}_{P}\) into uniformly sized annular sub-volumes \(\mathcal {I}_{N}^{j}\), where j is the number of uniformly-sized annular bands, such that \(j \in \{1,\dots ,k\}\), and k is an user-defined proximity parameter dependent on the distance g from the tumor margin. For each \(\mathcal {I}\), there exists a corresponding tumor segmentation \(\mathcal {T}\) and vessel segmentation \(\mathcal {V}\).
3.2 VaNgOGH Descriptor
-
1.
Segmentation and skeletonization of tumor and vasculature
A segmentation algorithm is applied to \(\mathcal {I}\), yielding a volume \(\mathcal {T}\) containing the tumor, and a volume \(\mathcal {V}\) containing the surrounding tumor-associated vasculature (Fig. 2(a)). \(\mathcal {T}\) is subtracted from \(\mathcal {V}\) to ensure that there are no residual tumor voxels within the segmented vasculature. A fast marching approach is employed to compute the centerlines of vessels within \(\mathcal {V}\), forming \(\mathcal {S}\): a series of points in 3-dimensional cartesian space comprising the medial axis skeleton of \(\mathcal {V}\) (Fig. 2(b)). Segmentation algorithms utilized for each dataset are described in greater detail in Sects. 4.3 and 4.4.
-
2.
Obtain 2-Dimensional vessel network representations in cartesian and spherical domains.
-
(a)
Cartesian domain: \(\mathcal {S}\) is projected along the plane of image acquisition, z, in order to obtain a 2-dimensional representation of the vasculature, \(\mathcal {V}_{xy}\), which depicts the vascular network in the XY plane (Fig. 2(c)).
-
(b)
Spherical domain: The vascular network is converted to spherical coordinates and projected along each spherical axis to yield three 2-D representations of 3-D vessel orientation with respect to the tumor centroid. Each point within \(\mathcal {S}\) is converted to its spherical coordinates relative to the tumor centroid, D. Let \(x_D\), \(y_D\), and \(z_D\) represent distance from D of a point \(S_i\) within S along the corresponding cartesian axes. Each \(S_i\) then corresponds to an azimuth \(\alpha \) and an elevation \(\phi \), indicating rotation around the z-axis and angle from the XY-plane, respectively, such that \(\alpha = \arctan ^(\frac{y_D}{x_D})\) and \(\phi = \arctan (\frac{z_D}{\sqrt{x_D^2 +y_D^2}})\). A third spherical coordinate, r, given by \(argmin(\sqrt{ (x_D - \mathcal {T}^j_x)^2 + (y_D - \mathcal {T}^j_y)^2 + (z_D - \mathcal {T}^j_z)^2}) \) defines the Euclidean distance between \(S_i\) and the nearest voxel within \(\mathcal {T}\). This conversion is repeated for each \(S_i\) in S, yielding a 3-D skeleton within the spherical coordinate space, \(\mathcal {S}_{r\alpha \phi }\). \(\mathcal {S}_{r\alpha \phi }\) is projected along each spherical dimension to yield the following 2-dimensional representations of 3-D vessel orientation relative to the tumor centroid in spherical space: \({V}_{r\alpha }^{j}\), azimuth angle with respect to Euclidean distance from the tumor (Fig. 2(f)); \({V}_{r\phi }^{j}\), elevation angle with respect to Euclidean distance from the tumor (Fig. 2(e)); and \({V}_{ \alpha \phi }^{j}\), elevation angle with respect to azimuth angle (Fig. 2(d)).
-
(a)
-
3
Sliding Hough transforms and aggregate peak orientations.
-
(a)
Computation and aggregation of localized Hough transforms in the cartesian space: Using a \(\mathcal {N}\times \mathcal {N}\) sliding window \(\mathcal {W}\) with an offset of k pixels, each pixel in \(\mathcal {V}_{xy}\) is mapped to an accumulator space using the Hough Transform, where the equation of a line is represented by \(y=(-\frac{cos \theta }{sin \theta })x + (\frac{\rho }{sin \theta })\). This transforms the spatial coordinate system (x, y) to the polar coordinate system \((\rho , \theta )\), such that for every point on the medial axis representation \(\mathcal {V}_{xy}\), there exists a unique sinusoid in the Hough accumulator space (Fig. 2(g)). The five grid locations accumulating the most sinusoid crossings are identified for each \(\mathcal {W}\). Feature set \(\mathcal {F}_{xy}\) then comprises the \(\theta \) values associated with the five most prominent peak orientations such that \(\mathcal {F}_{xy}\) = \([\theta _1, \theta _2, ... \theta _5]\).
-
(b)
Computation of localized Hough transforms on spherical projections within annular sub-volumes: For a given annular sub-volume outside the tumor \(\mathcal {I}_{N}^{j}\), 2-D spherical representations are obtained from vessels only within the sub-volume, denoted as \({V}_{r\alpha }^{j}\), \({V}_{r\phi }^{j}\), and \({V}_{ \alpha \phi }^{j}\). Peaks are computed from 2-D spherical representations using the above approach. This is repeated for all annular bands \(j \in \{1,\dots ,k\}\). Peak orientations are concatenated for all annular bands, yielding a single feature vector, i.e. \(\mathcal {F}_{r\alpha }\) = [\(\mathcal {F}_{r\alpha }^1\), \(\mathcal {F}_{r\alpha }^2\), ... \(\mathcal {F}_{r\alpha }^k\)], \(\mathcal {F}_{r\phi }\) = [\(\mathcal {F}_{r\phi }^1\), \(\mathcal {F}_{r\phi }^2\), ... \(\mathcal {F}_{r\phi }^k\)], and \(\mathcal {F}_{\alpha \phi }\) = [\(\mathcal {F}_{\alpha \phi }^1\), \(\mathcal {F}_{\alpha \phi }^2\), ... \(\mathcal {F}_{\alpha \phi }^k\)].
-
(a)
-
4.
Computation of VaNgOGH descriptor
The final VaNgOGH feature set, \(\mathcal {F}_{V}\), is a concatenation of the first order statistics, mean, median, standard deviation, skewness, and kurtosis, of \(\mathcal {F}_{xy}\), \(\mathcal {F}_{r\alpha }^k\), \(\mathcal {F}_{r\phi }^k\), and \(\mathcal {F}_{\alpha \phi }^k\)
4 Experimental Results and Discussion
4.1 Data Description
Dataset 1 included axial breast DCE-MRIs, collected prior to administration of neoadjuvant chemotherapy with a 1.5/3T magnet. Dataset 2 included non-contrast lung CT scans, collected from two sites. Patients were divided at random into training and testing cohorts as shown in Table 1. All studies were acquired as part of an Institutional Review Board-approved, HIPAA-compliant protocol.
4.2 Comparative Strategies and Classifier Construction
Intra- and Peri-tumoral Radiomics: For Experiment 1, VaNgOGH was compared against four radiomic features capable of predicting response on pre-treatment DCE-MRI, previously published in [6]. The feature comprised 2 intra-tumoral features (CoLlAGe Info1 [8], Laws S5R5) and 2 peri-tumoral features (Laws L5S5, CoLlAGe Entropy). In Experiment 2, we used the same list of features described in [6], with the most important features being three intranodular low frequency Gabor and a perinodular Laws W5E5 feature.
Convolutional Neural Network: We used a multi-layer 2D LeNet-like architecture [9], comprising three sets of convolutional, activation (ReLU), and pooling layers, followed by a fully-connected layer, activation, another fully-connected, and finally a softmax classifier. In this patch-based classification approach, the softmax classifier returns the probability of each patch belonging to the two classes of interest. The model was trained over 100 epochs.
Feature Selection and Classifier Construction: Once the VaNgOGH and other radiomic features were extracted, a set of 4 top features were selected by Wilcoxon rank-sum test and training of a linear discriminant analysis classifier was performed in a 3-fold cross-validation setting across 100 iterations. The locked-down model was then applied to the independent validation cohorts. Performance of individual classifiers was assessed by the area under the receiver operating characteristic curve (AUC) (Fig. 2(h)).
4.3 Experiment 1: Pre-treatment Response Prediction in Breast Cancer DCE-MRI
Implementation Details. Subtraction images were derived from MRI scans prior to, and immediately following contrast agent injection. Multi-scale vessel enhancement [10] was performed to emphasize the vasculature, which was then isolated by thresholding. A series of morphological operations were performed to remove noise and join adjacent vascular regions. Tumor boundaries were delineated by an expert radiologist. Hough transforms were applied to image projections using a sliding window size of \(\mathcal {N}\) = 30 pixels with a step size k = 5 pixels. Spherical projections were performed within annular sub-volumes with a radial width of 25 pixels out to a maximum radial distance of 100 pixels, with a step size = 12.5 pixels.
Results: pCR demonstrates a less chaotic vascular network, potentially evidenced by projection images in Fig. 1(c) on the cartesian plane and a distance-elevation image in Fig. 1(d) on the spherical plane. Hough transformation on (c) and (d) further accentuates the differences in vessel arrangement by detecting the orientation of straight line segments in the accumulator grid. The best discriminating VaNgOGH features comprised the standard deviation and kurtosis of \(\mathcal {F}_{\alpha \phi }^1\) and skewness and kurtosis of \(\mathcal {F}_{r\phi }^1\). Kurtosis of \(\mathcal {F}_{r\phi }^1\) was elevated in pCR (Fig. 1(e)), indicating a reduced disorder of vessel orientation in the \(\mathcal {V}_{r\phi }\) space. The training AUCs were .63 ± .06 and .64 ± .07 using VaNgOGH and texture features, respectively. Combining VaNgOGH and texture resulted in an improved testing AUC of .79. Testing AUCs have are shown in Fig. 3.
4.4 Experiment 2: Malignancy Diagnosis for Lung Nodules
Implementation Details. Distinguishing granulomas from adenocarcinomas is amongst the most challenging clinical problems for lung radiologists. Nodules were manually segmented by a cardiothoracic radiologist. To obtain the vasculature, lung regions were first isolated from the surrounding anatomy using a multi-threshold based algorithm [11]. This was followed by region growing [12]. The center of gravity of the segmented nodules was used as the initial seed point for the region growing algorithm [13]. Within the nodule volume, seed points were initialized at random locations. Based off the intensity similarity of the seed points and surrounding pixels, an initial region was iteratively grown to encompass the nodule and associated vasculature. VaNgOGH features were extracted using the approach described in Sect. 4.3.
Results: As may be observed from Fig. 4, although both nodules are highly vascularized, spherical projections (below) accentuate the elevated disorder of the vascular network in adenocarcinoma, whereas granuloma vessel orientations are predominantly linear in the \({V}_{ \alpha \phi }^{j}\) (d) and \({V}_{ r \alpha }^{j}\) (e) spaces. VaNgOGH successfully separated adenocarcinomas and granulomas with an AUC of .65 ± .06 in the training set. The top feature set included one statistic for each view (standard deviation of \(\mathcal {F}_{r\phi }^1\), median of \(\mathcal {F}_{\alpha \phi }^1\), kurtosis of \(\mathcal {F}_{xy}^1\), and skewness of \(\mathcal {F}_{r\alpha }^1\)). Figure 4 shows elevated vascular disorder in adeno, e.g. increased standard deviation of \(\mathcal {F}_{r\phi }^1\) in (d). Performance of VaNgOGH in the independent validation set was comparable to that of the CNN, as shown in Fig. 3.
5 Concluding Remarks
In this work, we presented a new radiomic descriptor, VaNgOGH, that quantifies disorder of the tumor-associated vascular network by assessing the morphology of vessel orientation across multiple spatial domains. To the best of our knowledge, VaNgOGH is the first radiomic descriptor of tumor-associated vascular morphology. We demonstrated the ability of VaNgOGH to (a) predict benefit of neoadjuvant chemotherapy in breast cancer patients on DCE-MRI (N = 76), and (b) distinguish malignant adenocarcinoma from visually confounding granuloma on lung CT (N = 81). VaNgOGH performed comparably to deep learning and state of the art radiomic approaches, and provides greater biological interpretability. When considered in conjunction with an established radiomic response signature in breast MRI, VaNgOGH further improved pCR identification. Future work will entail larger validation studies and also evaluation on other use cases.
References
Yamazaki, K., et al.: Tumor angiogenesis in human lung adenocarcinoma. Cancer 74, 2245–2250 (1994)
Grunewald, M., et al.: VEGF-induced adult neovascularization: recruitment, retention, and role of accessory cells. Cell 124, 175–189 (2006)
Bullitt, E., et al.: Tumor therapeutic response and vessel tortuosity: preliminary report in metastatic breast cancer. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI 2006. LNCS, vol. 4191, pp. 561–568. Springer, Heidelberg (2006). https://doi.org/10.1007/11866763_69
Aerts, H.J.W.L., et al.: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014)
Li, H., et al.: Quantitative MRI radiomics in the prediction of molecular classifications of BCA subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2, 16012 (2016)
Braman, N.M., et al.: Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 19, 57 (2017)
Zheng, Y., et al.: Parenchymal texture analysis in digital mammography: A fully automated pipeline for breast cancer risk assessment. Med Phys. 42, 4149–4160 (2015)
Prasanna, P., Tiwari, P., Madabhushi, A.: Co-occurrence of local anisotropic gradient orientations (CoLlAGe): distinguishing tumor confounders and molecular subtypes on MRI. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8675, pp. 73–80. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10443-0_10
LeCun, Y., et al.: Gradient-based learning applied to document recognition. IEEE 86, 2278–2324 (1998)
Frangi, A.F., Niessen, W.J., Vincken, K.L., Viergever, M.A.: Multiscale vessel enhancement filtering. In: Wells, W.M., Colchester, A., Delp, S. (eds.) MICCAI 1998. LNCS, vol. 1496, pp. 130–137. Springer, Heidelberg (1998). https://doi.org/10.1007/BFb0056195
Hu, S., et al.: Automatic lung segmentation for accurate quantitation of volumetric x-ray CT. IEEE TMI 20, 490–498 (2001)
Adams, R., et al.: Seeded region growing. IEEE PAMI 16, 641–647 (1994)
Sethian, J.A.: Fast marching methods. SIAM Rev. 41, 199–235 (1999)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Braman, N., Prasanna, P., Alilou, M., Beig, N., Madabhushi, A. (2018). Vascular Network Organization via Hough Transform (VaNgOGH): A Novel Radiomic Biomarker for Diagnosis and Treatment Response. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_89
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_89
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)