Abstract
The quantitative use of medical images often requires an intensity scaling with respect to the signal from a well-characterized anatomical region of interest. The choice of such a region often varies between studies which can substantially influence the quantification, resulting in study bias hampering objective findings which are detrimental to open science. This study outlines a list of criteria and a statistical ranking approach for identifying normalization region of interest. The proposed criteria include (i) associations between reference region and demographics such as age, (ii) diagnostic group differences in the reference region, (iii) correlation between reference and primary areas of interest, (iv) local variance in the reference region, and (v) longitudinal reproducibility of the target regions when normalized. The proposed approach has been used to establish an optimal normalization region of interest for the analysis of Quantitative Susceptibility Mapping (QSM) of Magnetic Resonance Imaging (MRI). This was achieved by using cross-sectional data from 119 subjects with normal cognition, mild cognitive impairment, and Alzheimer’s disease as well as and 19 healthy elderly individuals with longitudinal data. For the QSM application, we found that normalizing by the white matter regions not only satisfies the criteria but it also provides the best separation between clinical groups for deep brain nuclei target regions.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Emerging quantitative medical imaging techniques have become a promising research tool for investigating metabolic, functional, and molecular properties of tissues. An accurate quantification of neuroimaging data such as Magnetic Resonance Imaging (MRI), Computed Tomography (CT) or Positron Emission Tomography (PET), often requires intensity scaling or normalization. The intensity normalization aims at accounting for global variations between subjects as well as uncontrolled biological, experimental, and imaging factors that might otherwise bias the results. A ratio of the measured signal to a pre-defined reference region is one common normalization method. Indeed, various normalization regions have been utilized in amyloid PET [1, 2], 18F-FDG PET [3, 4], arterial spin labeling MRI [5,6,7], quantitative susceptibility mapping (QSM) [8,9,10,11,12] or dynamic susceptibility contrast [13]. An ideal reference region should be spared from pathology in the disorder under study, should have non-specific associations (saturable) to the imaging method and should have a stable measurement. Seldom can a consensus normalization region be established as it depends on the disease, cohort, age, and imaging parameters or tracers, resulting in unwanted variability in reported outcomes [6, 8, 9], even using the same imaging modality and/or pathological cohort.
Intensity normalization methods scale the image by computing the ratio of regions of interest (ROI) values to the average of all voxels within a reference region [1, 2, 5, 9] or a cluster of regions [14, 15]. Intensity normalization aims to reduce measurement variability caused by uncontrolled physiological differences among subjects and imaging techniques. The ideal approach for normalization depends on the underlying source of the variability. Using QSM as an example in this paper, the magnetic susceptibility (i.e. χ) is inferred from a dipole kernel operation, which, due to the zero value in the origin of the kernel in the Fourier domain (DC component), leads to an arbitrary baseline intensity shift in the reconstructed image [16]. Therefore, intensity normalization using a reference region is essential to address this global shift before reporting and comparing QSM findings. Previous studies reported the use of different reference regions including the cerebrospinal fluid (CSF) of the posterior ventricles [8, 11], posterior white-matter (WM) with lower variance across subjects [9], middle frontal WM [17], occipital WM [10], and whole brain or cortical grey-matter [12].
Previous studies established a series of objective criteria for a normalization region which included between- and intra-diagnostic-group differences [3, 11, 13, 17, 18] and associations with age and disease stage in a reference region [11, 17, 19]. However, these analyses were limited to the previously reported list of potential reference regions and the decision was made based on qualitative approaches.
Given the lack of consensus on the choice of the reference region, the existence of a range of imaging modalities and disorders, the current work aims to outline a general framework for identifying objectively a reference region throughout the brain. A series of quantitative criteria are proposed, which were evaluated for QSM-MRI modalities on a cross-sectional and longitudinal cohort of individuals with dementia and normal ageing. The outcome from each criterion was ranked and combined to produce a single rating metric for each region. A power analysis was performed to evaluate the impact of utilizing each reference region on detecting between diagnostic-group differences.
2 Method
2.1 Dataset
Two subsets of data based on the availability of cross-sectional and longitudinal QSM-MRI from the Australian Imaging Biomarkers and Lifestyle (AIBL) were included [20]. For the cross-sectional analysis, 119 QSM scans comprised of 69 cognitively normal (CN), 22 mild cognitive impairment (MCI) and 28 Alzheimer’s disease (AD). For longitudinal analysis, a baseline and 18-month follow-up QSM scans for 19 CN subjects without ApoE-ε4 alleles, Clinical Dementia Rating (CDR) = 0, Mini-Mental State Examination (MMSE) > 28 were included. Detailed demographic information is provided in Table 1.
2.2 Image Acquisition
MRI images were acquired on a 3T Siemens Tim Trio scanner with a 12-channel head coil. Subjects underwent anatomical T1-weighted (T1 W), gradient echo (GRE). The T1 W images were acquired using a standard 3D MPRAGE sequence with 1 × 1×1 mm3 resolution, TR/TE/TI = 1900/2.55/900 ms, flip angle 9°, field of view 256 × 256, and 160 slices. 3D GRE images used for QSM were acquired with 0.93 × 0.93 mm in-plane resolution and 1.75 mm slice thickness, repetition time/echo time of 27/20 ms, flip angle 20° and field of view 240 × 256, and 80 slices.
2.3 Image Post-processing
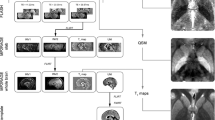
T1-weighted MPRAGE data were segmented into 73 grey-matter (GM) and 32 white-matter (WM) regions by segmentation propagation of an atlas database which had been previously parcellated using the Neuromorphometrics (63 subjects) [21] and FreeSurfer (FS) WM parcellation [22], respectively. The Neuromorphometrics and FS parcellations were then refined by the CSF, GM and WM segmentations obtained using the expectation maximization segmentation algorithm [23] directly applied to the T1-weighted images.
In order to reconstruct QSM images, phase offsets between each channel of the coil were removed by weighting the magnitude of the corresponding channel, and then combined to form a single-phase image. STI Suite software (version 2.2) was used for QSM dipole inversion process [24].
2.4 Reference Region Analysis
For this analysis, 100 reference regions including whole brain, CSF, WM, GM and 65 sub-regions from GM and 31 sub-regions from WM were used. As primary regions neocortex, frontal, parietal, temporal, occipital and hippocampus were considered. For each reference or primary region, a mean value for QSM was computed. To be able to combine statistical models, age and the regional mean values were scaled to zero mean and unit variance. A list of proposed criteria to identify a reliable reference region is as follows:
-
C1. Reference region association with subject demographics such as age and sex: there should be no or minimal association. This criterion was tested based on the β coefficient and standard error (SE) from a linear model with reference region as the dependent variable and subject demographics as covariates (lower β and SE are preferred for each covariate).
-
C2. Reference region diagnostic-group separation effect (e.g. between CN and MCI/AD participants): there should be no or minimal group differences. Any significant group effect in the reference region, e.g. MCI vs. AD, could impose a group difference when looking at the primary regions and therefore bias the normalized values. This was tested based on the β coefficient and SE from a linear model with reference region as the dependent variable, and diagnostic groups (CN, MCI and AD) as a covariate (lower β and SE are preferred for each covariate).
-
C3. Reference region correlation with other primary areas of interest e.g. cortical/subcortical regions: there should be a positive or negative association. The undesired change in the image is a global effect meaning that the measured values (both in the reference and target regions) are dependent. This was tested based on the β coefficient and SE from a linear model with the primary region as the dependent variable and reference region as independent variable while adjusting for age and gender (higher β and lower SE are preferred for the independent variable).
-
C4. Reference region signal variations: there should be a minimum variance. A lower variance in the reference region (of both CN and MCI/AD participants) shows the stability of the measurement as being unaffected by the pathology or a minimal contamination from neighboring structures (e.g. vessels) as well as other physiological or uncontrolled study confounds. This was tested by computing coefficient of variation (CV) where CV = standard deviation/mean (lower CV value is preferred).
-
C5. Longitudinal reproducibility of the primary region when normalized: there should be an improved reproducibility. Linear mixed models were used to assess the relationship between reference and primary regions over the two-time points controlled for age and gender modelled as QSM ~ Age*Gender + Region/Time + (1|ID). A lower β and SE are preferred for the Time (measured in years/months) by region interaction indicating little change between the reference region and the region of interest over time.
To combine all the criteria and obtain a single value representing the goodness-of-reference for normalization, the outcome of each criterion was ranked to 0 to 1 (higher the better) and then averaged to create a composite score. The Cohen’s D metric was used as well to compute the overall performance of each reference region in terms of improving the diagnostic accuracy (MCI and AD compared to CN) as previously employed by [3, 4, 19].
3 Results
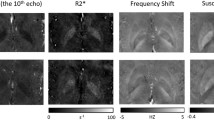
The top 5 reference regions (out of 100), when used for normalizing the primary regions of interest, are shown in Table 2. The average SE for reference regions (not shown here) were very similar and not significantly different. A comparison of previously reported reference regions and proposed top-ranked regions from Table 2 (first row) is shown in Fig. 1.
The Middle Frontal White Matter area had the best composite scores and ranked first overall from all the 100 areas considered for the normalization of QSM to study Alzheimer’s disease.
4 Discussion
This study proposed an objective framework for identifying a suitable reference region for intensity normalization with the purpose of removing global uncontrolled variations in quantitative images.
We argue in this paper that the selection of a normalization region should be done using objective and open criteria. This would allow fairer comparison between reported results removing doubts about introducing unwanted bias in the processing and interpretation. Furthermore, we also argue that the normalization regions should be dependent on the data acquisition and application. In our example, to study Alzheimer’s disease, it would be important that the normalization region does not introduce group difference, and the area that ranked first would be different if another target region (than the neocortex) was under investigation. However, our study shows that one reference region (i.e. Mid Frontal WM) ranked highly overall when investigating multiple brain regions.
Our method is a data-driven approach and as such results ought to be investigated, to avoid spurious selection that would have no justification for the application at hand. Indeed, in our QSM application, the orbital gyrus GM had a better effect size improvement to separate AD from CN, however, in terms of pathophysiology and its lower score for the other criteria do not support it as a suitable choice. This highlights the benefit of having a set of criteria instead of investigating only one (e.g. effect size in this case).
5 Conclusion
Normalization is essential for removing global modulation effects, and consequently improving the sensitivity of quantitative imaging in detecting disease-specific differences. This study proposed a systematic approach to objectively identify a suitable reference region and evaluated it on QSM-MRI data to identify the Mid Frontal WM as the best normalization area for investigating neocortical QSM signal.
References
Dore, V., et al.: CapAIBL: automated reporting of cortical PET quantification without need of MRI on brain surface using a patch-based method. In: Wu, G., Coupé, P., Zhan, Y., Munsell, B.C., Rueckert, D. (eds.) Patch-MI 2016. LNCS, vol. 9993, pp. 109–116. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-47118-1_14
Brendel, M., et al.: Improved longitudinal [18F]-AV45 amyloid PET by white matter reference and VOI-based partial volume effect correction. NeuroImage 108, 450–459 (2015)
Yakushev, I., et al.: Choice of reference area in studies of Alzheimer’s disease using positron emission tomography with fluorodeoxyglucose-F18. Psychiatr. Res. Neuroimaging 164(2), 143–153 (2008)
Dukart, J., et al.: Differential effects of global and cerebellar normalization on detection and differentiation of dementia in FDG-PET studies. NeuroImage 49(2), 1490–1495 (2010)
Mattsson, N., et al.: Association of brain amyloid-β with cerebral perfusion and structure in Alzheimer’s disease and mild cognitive impairment. Brain 137, 1550–1561 (2014)
Alsop, D.C., Casement, M., de Bazelaire, C., Fong, T., Press, D.Z.: Hippocampal hyperperfusion in Alzheimer’s disease. NeuroImage 42(4), 1267–1274 (2008)
Alsop, D.C., Detre, J.A., Grossman, M.: Assessment of cerebral blood flow in Alzheimer’s disease by spin-labeled magnetic resonance imaging. Ann. Neurol. 47(1), 93–100 (2000)
Acosta-Cabronero, J., Williams, G.B., Cardenas-Blanco, A., Arnold, R.J., Lupson, V., Nestor, P.J.: In vivo quantitative susceptibility mapping (QSM) in Alzheimer’s disease. PLoS ONE 8(11), e81093 (2013)
Acosta-Cabronero, J., Betts, M.J., Cardenas-Blanco, A., Yang, S., Nestor, P.J.: In vivo MRI mapping of brain iron deposition across the adult lifespan. J. Neurosci. 36(2), 364–374 (2016)
Langkammer, C., et al.: Quantitative susceptibility mapping (QSM) as a means to measure brain iron? a post mortem validation study. Neuroimage 62(3–2), 1593–1599 (2012)
Straub, S., et al.: Suitable reference tissues for quantitative susceptibility mapping of the brain. Magn. Reson. Med. 78(1), 204–214 (2017)
Feng, X., Deistung, A., Reichenbach, J.R.: Quantitative susceptibility mapping (QSM) and R2* in the human brain at 3T: evaluation of intra-scanner repeatability. Z. Med. Phys. 28(1), 36–48 (2018)
Lacalle-Aurioles, M., et al.: Is the cerebellum the optimal reference region for intensity normalization of perfusion MR studies in early Alzheimer’s disease? PLoS ONE 8(12), e81548 (2013)
Borghammer, P., Aanerud, J., Gjedde, A.: Data-driven intensity normalization of PET group comparison studies is superior to global mean normalization. NeuroImage 46(4), 981–988 (2009)
Dukart, J., et al.: Reference cluster normalization improves detection of frontotemporal lobar degeneration by means of FDG-PET. PLoS ONE 8(2), e55415 (2013)
Koch, K.M., Papademetris, X., Rothman, D.L., de Graaf, R.A.: Rapid calculations of susceptibility-induced magnetostatic field perturbations for in vivo magnetic resonance. Phys. Med. Biol. 51(24), 6381–6402 (2006)
Fazlollahi, A., et al.: A normalisation framework for quantitative brain imaging; application to quantitative susceptibility mapping. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), pp. 97–100 (2017)
Schwarz, C.G., et al.: Optimizing PiB-PET SUVR change-over-time measurement by a large-scale analysis of longitudinal reliability, plausibility, separability, and correlation with MMSE. NeuroImage 144, 113–127 (2017)
Yakushev, I., et al.: SPM-based count normalization provides excellent discrimination of mild Alzheimer’s disease and amnestic mild cognitive impairment from healthy aging. NeuroImage 44(1), 43–50 (2009)
Ellis, K.A., et al.: The Australian imaging, biomarkers and lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int. Psychogeriatr. 21(4), 672–687 (2009)
Tzourio-Mazoyer, N., et al.: Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15(1), 273–289 (2002)
Salat, D.H., et al.: Regional white matter volume differences in nondemented aging and Alzheimer’s disease. NeuroImage 44(4), 1247–1258 (2009)
Leemput, K.V., Maes, F., Vandermeulen, D., Suetens, P.: Automated model-based tissue classification of MR images of the brain. IEEE Trans. Med. Imaging 18(10), 897–908 (1999)
Li, W., Avram, A.V., Wu, B., Xiao, X., Liu, C.: Integrated Laplacian-based phase unwrapping and background phase removal for quantitative susceptibility mapping. NMR Biomed. 27(2), 219–227 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Fazlollahi, A. et al. (2018). A Framework to Objectively Identify Reference Regions for Normalizing Quantitative Imaging. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-00928-1_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00927-4
Online ISBN: 978-3-030-00928-1
eBook Packages: Computer ScienceComputer Science (R0)