Abstract
This chapter will present the computational approach of registering images from different modalities based on manual selection of matching pairs of landmarks. Here we will present an image registration workflow based on MATLAB’s image processing toolbox using the identification of sites of clathrin-mediated endocytosis by correlative light electron microscopy (CLEM) as an example. In the Appendix section, we will discuss the concept of image transformations and how to generate them based on pairs of landmarks. We will also learn how to fit a 2D Gaussian for a more accurate positioning of the landmarks.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
This chapter will present the computational approach of registering images from different modalities based on manual selection of matching pairs of landmarks. Here we will present an image registration workflow based on MATLAB’s image processing toolbox using the identification of sites of clathrin-mediated endocytosis by correlative light electron microscopy (CLEM) as an example. In the Appendix section, we will discuss the concept of image transformations and how to generate them based on pairs of landmarks. We will also learn how to fit a 2D Gaussian for a more accurate positioning of the landmarks.
6.1 Introduction
The purpose is to use Fluorescence Microscopy (FM) to localize clathrin vesicles, and to correlate it with Electron Microscopy (EM) to identify their ultrastructure (◘ Fig. 6.1), based on the use of beads, as it was done in Avinoam et al. (2015). We will introduce the basic concept of image registration and dedicated MATLAB image processing commands to register light microscopy images of clathrin-mediated endocytosis and corresponding electron microscopy images to reveal the underlying ultrastructure (Avinoam et al. 2015). We will also discuss how enhancing the localization accuracy of fluorescence signals will improve the registration accuracy.
Clathrin in conjunction with other proteins involved in endocytosis forms a lattice that can dramatically change the shape of the plasma membrane to form a vesicle. Top row: electron microscopy (EM) image. Bottom row: schematic of top row, with the plasma membrane in black and clathrin and associated proteins in red. (Image provided by Ori Avinoam, EMBL and Weizmann Institute of science, from the data published in Avinoam et al. 2015)
The first task in a typical CLEM experiment is to identify the two image datasets to be registered. The data from the second image modality (in the CLEM case this is EM) will likely be acquired in a targeted approach using the previous light microscopy observations. For a good review about different approaches of targeting the same area, the reader can be referred to de Boer et al. (2015).
6.2 Data Presentation
All data used are available using the DOI:
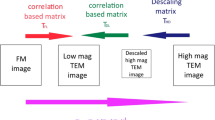
The data we will use here were acquired using the protocol described in Avinoam et al. (2015). In order to obtain a higher-resolution insight into the ultrastructure underlying a fluorescence signal, we acquired images at high magnification (pixel size ≈ 1 nm, FOV 2 μm). The field of view at this magnification however is too small to contain enough landmark beads for a direct registration of the FM data. Therefore we need to first register to a lower magnification EM overview and then to the final high-resolution EM data (see ◘ Figs. 6.2 and 6.6 for an overview of datasets and scales).
We will register the fluorescent image ex1_FM.tif on the EM data ex1_EM.tif using beads visible in both modalities. We will then register ex1_EM.tif on ex1_highmag.tif which is acquired at the same depth of the sample using black gold particles. The position of interest showing clathrin is deeper in the cell, and appears as the other non black slice in this figure in the high mag EM stack. It is named ex1_highmag_poi.tif in this chapter
Important note: on EM images the polystyrene beads appear as extended gray circles, not as black spots. The small black spots spread across the EM image are gold particles used to register EM data on itself for tilt correction (tomographic reconstruction) or alignment. We will use these gold beads as landmarks to accurately register the EM images of different magnifications.
The MATLAB functions described here will all handle 2D image data. Therefore we need to reduce the EM source data from the tomographic volumes. We can either choose single slices or an average of a small subset (5–10) slices from the source volumes. This can easily be done in Fiji (Schindelin et al. 2012), Icy (de Chaumont et al. 2012) or similar software. Here we could also adjust the contrast/brightness of the images to facilitate recognition of landmark features later in the process.
We already have prepared the matching images from FM and EM and made them available in the data directory. Ideally, we could now run an automated routine that would provide us with appropriate landmark features in both images. However, as the image data from FM and EM are intrinsically complementary and thus very different, in most of the cases the landmarks need to be selected manually.
We can find the respective preprocessed files for the 2D registration work flow in the data directory:
-
FM data: multi-color wide-field fluorescence acquisition in these channels: (red: RFP, green: GFP, blue: 380 nm emission for beads) ex1_FM.tif
-
EM data low-magnification: we usually acquire a tomographic tilt series of images that after reconstruction results in a three-dimensional image stack. As the beads are located on top of the specimen, they can be identified in this stack. Usually we use an average of about 5 slices to have the best signature of the beads for the 2D registration. ex1_EM.tif
-
EM data high-magnification: this comes from a high-resolution tomographic stack. For the 2D registration, we use a single slice that resembles the section selected for the low-mag registration, at the top of the tomogramm where the beads are. ex1_highmag.tif
-
EM data high-magnification point of interest: this is a single slice from the same tomogramm that contains the structure of interest (here potential clathrin vesicles). ex1_highmag_poi.tif
An example of expected result navigating between these scales and images is shown on ◘ Fig. 6.9, where the red fluorescent spots are associated with specific ultrastructures.
6.3 Overview of Data Processing
-
Step 1: Read and display EM and FM images
-
Step 2: Manually identify landmarks pairs
-
Step 3: Refine localisation by Gaussian fitting of the landmarks beads localization
-
Step 4: Compute the rigid transform + scale from the list of paired landmarks localization.
-
Step 5: Apply the Transformation (and discuss interpolation and potential artifacts)
-
Step 6: Evaluate the confidence in structure matching
6.4 Tools Description
All codes are available here:
We will use the commercial software package MATLAB as well as its Image Processing and Optimization toolboxes (alternatively to the optimization toolbox, we can use the CurveFitting toobox; both versions of the code are provided). MATLAB provides a set of registration tools, gathered under the topic “image registration” in the MATLAB documentation (mat).
In particular, we will use:
-
cpselect: built-in function from MATLAB Image Processing Toolbox allowing to provide a user interface for the selection of landmarks pairs.These are called control points in MATLAB language.
-
fitgeotrans: built-in function from MATLAB Image processing toolbox allowing to fit a defined geometric transform matching pairs of landmarks (control points in Matlab language).
To refine the localisation of fluorescent spots correponding to the landmark beads, we will fit a 2D Gaussian to the beads’ signal in a cropped image (see ◘ Fig. 6.11). For this procedure, there are two possible toolboxes in MATLAB: the Curve Fitting Toolbox, or the Optimization Toolbox. In particular, we will use:
-
if we use the curve fitting toolbox we can use fit: built-in function from MATLAB Curve Fitting Toolbox allowing to find the parameters of a function which best fits given data. The advantage is that it provides also confidence intervals in fitting.
-
if we use the optimization toolbox we can use lsqnonlin: built-in function from MATLAB Optimization Toolbox allowing to find the parameters of a function which best fits given data by least square fitting.
In our case, the function will be a 2D Gaussian equation and the data will be the pixels values of the cropped image around the manually selected landmarks.
As usual with Matlab, after downloading the code and data, we need to update our matlab path to include the code directory. The code directory contains files with the cascade of code used, as a correction or catch-up hint. It also contains the 2D Gaussian fitting functions.
6.5 Application to a CLEM Experiment
6.5.1 CLEM Workflow Overview and Preparation
The procedure of registering light microscopy data to electron microscopy data requires the landmarks to be clearly visible in both imaging modalities. We found fluorescently labelled polystyrene beads to match these criteria best (Kukulski et al. 2011). We do not want their signal to interfere with the fluorescence signal of interest, therefore the beads need to fluoresce in another channel. When using beads that only fluoresce in a different channel, shifts between the channels due to optical aberrations or stage instabilities during the acquisition will deteriorate the registration accuracy of our signal of interest. We either need to correct for these shifts or choose fluorescence beads that are both visible in the channel of interest and in another channel in order to be able to distinguish them from the real feature we want to localize. This is the case for our test dataset (Avinoam et al. 2015). The beads will be our landmarks (or control points in Matlab language). The typical feature of interest, an intracellular structural or morphological feature would have a size of about 100 nm. This is way beyond the diffraction limit of conventional fluorescence microscopy. The typical pixel size of FM data is on the order of 80–100 nm, so a single pixel difference in localization in the FM image could distort the registration of our feature by 100%. Therefore it is necessary to perform a precise sub-pixel localization not only on the fluorescent signal of interest but also on those of the landmarks.
The workflow to register the light microscopy data onto the EM data will be the following:
-
identify the area of interest based on the target fluorescence signal (step 1)
-
identify the locations of the surrounding fluorescent beads using the different channels
-
identify the location of the beads visible in the EM image
-
mark the matching landmark pairs in both images (step 2)
-
precisely determine their localization in the FM image (step 3)
-
calculate the image transformation (step 4)
-
evaluate the confidence in structure matching (step 5)
-
mark the coordinate of the feature of interest
-
precisely determine its localization in the FM image
-
apply the coordinate transformation
-
create the output data (image overlays, coordinate lists, …)
The field of view in which we observe the EM features is not sufficiently large to capture enough landmark beads. Therefore, we need to perform an initial registration of the FM data with a lower-magnification overview and then a second registration to the higher magnification EM data. This second registration can be done in an automated fashion, as it is basically just a change in magnification and thus the image features are the same.
6.5.2 Labeling of Landmark Pairs
MATLAB’s Image Processing Toolbox offers a selection of graphical tools to mark positions in an image. The command to mark coordinates in a displayed image is be ginput. However, we would like to assign coordinate pairs in the two image modes simultaneously. Therefore, the tool of choice is cpselect. This function expects the two images to be displayed as input and will give us two coordinate lists as a result of the point selection. The first image and associated coordinates are referred-to as “moving” and the second as “fixed”. This means that the first image is the one whose coordinate system will be transferred onto the second. Note: Moving image is also sometimes called source image, and fixed image called target image.
6.5.2.1 Correlation from Low Magnification Tomogram to High Magnification EM Image
In order to locate the feature of interest in or high-resolution dataset, we need to register the low-magnification EM data, where we will map the FM data onto, to the high-magnification images. We can do this using the exact same tools as for registering the FM data with the low-magnification EM data. We will do this procedure first to get familiar with the tools, as we have very similar features in both images. We will use the gold beads (black spots) present on the specimen as common landmarks to register the EM images of different resolutions (◘ Fig. 6.3).
The manual selection process of landmark pairs in MATLAB’s cpselect tool showing EM images at different magnifications. The “Add point and Predict Match” tool is selected and the predicted position for point 4 is indicated in the right panel in yellow
The manual selection process of landmark pairs in light (right) and electron microscopy (left) images using MATLAB’s cpselect tool
Illustration of the relative size of EM and FM images. The EM images (low mag and high mag, size: 2048 × 2048 pixel) are overlaid for comparison of scale, after registration. Red: red channel (showing clathrin and beads) from FM, Gray: low mag EM. Yellow: High Mag EM after registration on the low mag EM
First, we want to load our images. em is the overview (low-magnification) EM image, hm is the high-magnification image at the same z-height that also contains the gold beads we will use as landmarks. The slice of interest that contains the ultrastructure is also loaded (sm).
1 hm = imread(’ex1_highmag_1.tif’); 2 sm = imread(’ex1_highmag_poi.tif’); 3 em = imread(’ex1_EM.tif’);
We can display either of the images using the imshow command.
1 imshow(sm);
Now, we can open the two images next to each other in cpselect. The coordinates of landmark beads from the low-magnification image will be stored in c_lm, and those from the high-magnification EM image in c_hm. We have to make sure that we always click the corresponding landmark pairs in an alternating fashion between the two images to keep their correct association. The UI offers the option to use the already determined pairs (> 2) for a prediction of the corresponding point for each new clicked spot. Simply activate the second “Add point and Predict Match” toggle button on the top left of the images (◘ Fig. 6.4). Because we would like to store the clicked coordinates in two variables directly, we need to provide the ’Wait’,true option to cpselect to prevent MATLAB from running other processes while we pick the coordinates.
The final result of the image registration (left and center: ‘montage’ parameter) together with the overlay using the ‘blend’ option (right) for imshowpair
1 [c_hm,c_lm] = cpselect(hm,em,’Wait’,true);
Once done, simply close the cpselect window. The two output variables c_hm and c_lm contain the two coordinate lists of the landmark pairs that we need to generate our transformation.
We will generate the transformation that best relates these two sets of coordinates in the ► Sect. 6.5.5.
Now that we have seen the cpselect tool, we can use it to defind the landmark pairs for aligning the light-microscopy data to the low-magnification EM data.
First, we want to load our images. fm is the fluorescence image (FM), em will be the overview image from EM.
1 fm = imread(’ex1_FM.tif’);
We can display either of the images using the imshow command and automatically adjust the image contrast with imadjust. The FM image is a 16 bit 3-channels image. In order to work with it using the described Matlab tools, we need to choose a single channel to work with. In order to avoid chromatic aberrations in the registration process, and as the beads are visible in this channel, we use the channel of interest (Red, i.e. first channel) to mark the beads. Note that the blue channel (channel 3) contains only the beads, and could be used later one to differentiate beads and Clathrin.
1 fm1 = fm(:,:,1); % select red channel 2 imshow(imadjust(fm1));
Open the two images next to each other in cpselect. The coordinates of landmark beads from the FM image will be stored in c_fm, those from the EM image in c_em. We need to make sure that we always click the corresponding landmark pairs in an alternating fashion between the two images to keep their correct association. The UI offers the option to use the already determined pairs (>2) for a prediction of the corresponding point for each new clicked spot. Simply activate the second “Add point and Predict Match” toggle button on the top left of the images to use this option.
1 [c_fm,c_em] = cpselect(em,imadjust(fm1),’Wait’,true);
Because the polystyrene beads we use as landmark markers are very difficult to identify on the EM data (position of beads in EM data are shown in ◘ Fig. 6.10, some initialization points are provided for the ease of this demonstration. Let’s load their coordinates (this file is in the code directory):
1 load(’preselectedpoints.mat’);
This mat-file contains two variables:
em_cp_preselected and fm_cp_preselected.
1 [c_fm,c_em] = cpselect(em,imadjust(fm1),em_cp_preselected,fm_cp_preselected,’Wait’,true);
The cpselect window should now look like on ◘ Fig. 6.5.
The color overlay produced by imshowpair allows to check successful registration of the high-mag image to the low-mag
Try to add a point using the “Add point and Predict Match” toggle button.
Once we are done with selecting landmark pairs, close the cpselect window. The two output variables c_fm and c_em contain the two coordinate lists, in pixels, that we need in order to generate our transformation.
6.5.3 Generating the Transformation
In the case of a similarity, we would like to solve for the transformation matrix T in the system of equations described in equation 6.14. The lists of coordinates now consist of row vectors instead of the required column vectors, so we need to transpose them before the calculation:
1 T = c_em’ / [c_fm’;ones(1,length(c_fm))];
with the result giving us a transformation matrix like this:
1 T = 2 3 1.0e+03 * 4 5 0.0125 -0.0038 -5.2820 6 0.0040 0.0125 -9.1019
If we compare the (1,2) and (2,1) entries of the matrix, i.e. 0.0040 and −0.0038 (the coefficient b from Eq. 6.10), we notice that the solution does not exactly fulfill the prerequisites of a similarity (same magnitude in scaling in both axes). Let’s check what the transformation matrix looks like that we generate with fitgeotrans:
1 structT = fitgeotrans(c_fm,c_em,’similarity’); 2 T = structT.T’; 3 4 T = 5 6 1.0e+03 * 7 8 0.0124 -0.0040 -5.1138 9 0.0040 0.0124 -9.0235 10 0 0 0.0010
This matrix resembles the one generated before, but now describes a true similarity.
6.5.4 Applying the Transformation to Image and Coordinate Data
6.5.4.1 Transforming Images
We now would like to apply the transformation to find out where our fluorescent signal of interest is located within the EM image/volume. In order to transform an image we can use the MATLAB function imwarp. Obviously our initial FM image covers a much larger field of view than the EM image (◘ Fig. 6.6). We therefore need to provide the function with the scale and dimension of the target image. This is done using imref2D.
In order to generate the pair of registered images, we will now apply the transformation in the structured variable strucT computed by fitgeotrans, using imwarp.
1 em_geom = imref2d(size(em)); 2 fm_trans = imwarp(fm1,structT,’OutputView’,em_geom); 3 figure(1);imshowpair(em,fm_trans,’montage’); 4 %shows the images side-by-side 5 figure(2); imshowpair(em,fm_trans,’blend’); 6 %shows them merged
The result of these commands is shown in ◘ Fig. 6.7 (for one channel only).
Left: Clathrin signal identified by FM (green circles), note the bright signal of a polystyrene bead on the right; Middle: High mag EM top slice showing the gold beads and the polystyrene bead (right) on the surface of the specimen; Right: High mag EM slice some nm below in the tomogram showing the signal originating from complete vesicles (left, right) and from a forming invagination (center). This panel is generated by the example code
6.5.4.2 Transforming Coordinates
In order to accurately transfer a set of coordinates we can use ginput on the fluorescence image to select points of interest on the FM image. Several points can be entered. Do not forget to press the return key when done to quit ginput mode. We then transform the obtained coordinate list using transformPointsForward and display the resulting coordinates on top of the EM image.
1 figure(3); 2 imshow(imadjust(fm1)); 3 [x,y] = ginput; 4 [u,v] = transformPointsForward(structT,x,y); 5 figure(2), hold on, plot(u,v, ’*r’);
6.5.5 Registering the Low-Magnification and the High-Magnification EM Data
Now that we have transformed the pixel coordinates of the feature(s) of interest from the FM image onto the low-magnification EM image, we need to perform a second transformation in order to map these coordinates to the high-magnification data. Let’s use the landmark lists that we have generated in the beginning (of ► Sect. 6.5.2) and calculate the second registration between low and high EM magnifications.
1 lm2hm = fitgeotrans(c_lm,c_hm,’similarity’);
In order to test whether the transformation is correct, we would like to display the high-mag image in the context of the lower-mag overview. Therefore we need to warp it using the inverse transform.
1 hm2lm = invert(lm2hm); 2 hm_trans = imwarp(hm,hm2lm,’OutputView’,em_geom); 3 imshowpair(em,hm_trans);
The function imshowpair without options will display the result of the registration in a color overlay (◘ Fig. 6.8).
Example of correction of manual selection based on Gaussian fitting. The mesh is the Gaussian fit, the red dots are the original pixel intensities in the cropped image (corrected by their base value), the blue star is the original clicked position, the green star is the corrected position. Note that here the clicked values were already very good, but remember that 1 pixel correction is about 100 nm correction, i.e. about 100 pixels in the high magnification EM image
Let’s now apply the transformation to the high-magnification data to our target coordinates of the features of interest. We have already found their coordinates in the low-mag EM (u,v), so we need to apply our transformation lm2hm to these.
1 [x_final,y_final] = transformPointsForward(lm2hm,u,v); 2 figure;imshow(sm);hold all 3 scatter(x_final,y_final,100, ’go’); 4 %this code generates the lowest panel in the results figure
The scatter function here will draw green circles on top of the existing figure (high-mag EM) at the coordinates of the transformed positions (◘ Fig. 6.9).
Exercise: Try to re-create the top panel of ◘ Fig. 6.9.
6.6 Accuracy Estimation and Improvements
With the transformation matrix we obtained, we can calculate the transformed coordinates of our landmarks from FM to EM and compare them with the clicked positions.
1 c_em1 = T * [c_fm’;ones(1,length(c_fm))]; 2 % identical alternative: c_em1 = transfromPointsForward(a,c_fm); 3 figure; imshow(em); 4 hold all; 5 scatter(c_em(:,1),c_em(:,2),70, ’r+’); 6 scatter(c_em1(1,:),c_em1(2,:),70, ’bo’); 7 % determine the deviation for the predicted points and get average and standard deviation 8 pos_diff = c_em1(1:2,:)’ - c_em; 9 [mu,sig] = normfit(pos_diff);
This will give us an idea about the deviation of the landmark positions from their predictions in EM pixels (◘ Fig. 6.10). Optionally, we can try different types of transformations and compare the resulting deviations.
It is advised to check what happens to the matrix when the control points are moved to slightly different positions (using cpselect).
In the example we skipped step 3, the refining of FM coordinates using a Gaussian fit. In order to improve the accuracy of the registration, both the localization of the fluorescence signal of interest and of the landmark beads can be improved using a fit of the peak with sub-pixel accuracy. This fit can be performed directly in MATLAB during the workflow. We are going to add this step, just after the manual selection, for the FM fluorescent signal of the beads (Step 6.5.2).
For this we can use the provided script called GaussianFit_... that corresponds to the Toolbox available on our computer (either optimization or curve fitting Matlab toolbox). We should start by removing the suffix of the file needed such that it is called GaussianFit.m alone. To know which toolbox is available, type ver. The principle is the following:
-
Input parameters are the original FM image (fm1), the list of selected control points (or landmarks) on FM (c_fm), and a parameter in pixels that will give the crop size, called N.
-
For each control point
-
Crop the original image around the control point position, plus and minus N.
-
Try to fit a 2D Gaussian (we are assuming that the Gaussian is symmetric, i.e σ x = σ y = σ)
$$ G\left(x,y\right)=A{e}^{-\left(\frac{{\left(x-{x}_0\right)}^2+{\left(y-{y}_0\right)}^2}{2{\sigma}^2}\right)}\kern1.00em $$(6.1) -
Visually check the quality of the fit in a plot.
-
The parameters of this Gaussian will give us: A the amplitude of the Gaussian (peak height), σ its width, and (x 0, y 0) the central point of the Gaussian (peak position). Here we do not really care about the first two parameters, but (x 0, y 0) will allow us to correct the original position of the control point (x, y). Note that in a more advanced script, A and σ could be used for discriminating bad fitting automatically.
-
After having read the script, run it. N = 5 pixels will create a crop area of 11 × 11, which should be sufficient in our case.
1 corrected_positions = GaussianFit(fm, c_fm, 5);
It should display the fitting and the original data as shown in ◘ Fig. 6.11. Press any key to process the next control point (this is achieved by a pause command in the MATLAB script).
Check the results visually with cpselect, now showing the corrected positions on top of the image.
1 [c_fm,c_em] = cpselect(imadjust(fm1),em,corrected_positions,c_em, ’Wait’,true);
Exercise: Complete SimpleCLEMworkfow.m by placing the accuracy refinement by Gaussian fitting. Add also a plot of the relative position of FM control points to matching control points in EM: it can indicates a bias such as a small drift if it is not centered around 0. We can use normfit to study their distribution. One could also add an histogram of the distance by using hist. Solution is provided in CLEMworkflowwithstep3andsimpleerrorstudy.m.
Take Home Message
In this chapter we have shown the basic principle of post-acquisition coordinate registration. We have seen how to express linear transforms with matrices, and how to compute them from pair of coordinates. We have registered the FM image showing polystyrene beads to an EM image at a low magnification, with enough beads visible in the field of view. We have then registered the low magnification EM to an higher magnification EM. Transforms can then be combined by simply multiplying them to position the fluorescence image on the high magnification EM image. The code as described here was used for a number of CLEM studies in the recent years (Kukulski et al. 2012; Schellenberger et al. 2014; Avinoam et al. 2015; Hampoelz et al. 2016; Curwin et al. 2016), but accuracy estimation was computed using another more effective approach than in this module. In this chapter, the localization error was computed only for the control points. Investigating only the error for the predicted landmark coordinates (as in 6.6) leads to an underestimation of the error for the points of interest. These might be located distant to landmarks and therefore behave differently under the transformation.
The accuracy of a registration also strongly depends on the accuracy of the localization of the landmarks. The Gaussian fitting can help to to reduce the resulting error. When an accurate localisation is not possible, another way of reducing registration inaccuracy is to increase the number of landmark points, and to make sure that they surround the point of interest (see Paul-Gilloteaux et al. (2017) for a theoretical description of accuracy in registration).
The registration workflow demonstrated in this chapter is applied in 2D, but the 3D workflow can be constructed in a similar way by adding the z dimension to both the coordinates and the transformation matrix.
There are alternative tools that perform similar tasks include ec-clem (Paul-Gilloteaux et al. 2017), which also supports 3D registration and propose some automatic registration options, as well as an estimation of error in any points of the image. In addition, it can help selecting the type of transformation needed, in particular it can automatically detect if a elastic transformation is needed, e.g. if the sample underwent deformations due to the fixation process for example. As it was demonstrated in Paul-Gilloteaux et al. (2017), selecting an elastic transformation when not needed will actually augment the error in other points of the images than the landmarks (◘ Fig. 6.12).
The source image we will use to demonstrate the transformations. Microscope pictograph adapted from ant (2013)
Bibliography
Antibiotic Resistance Threats in the United States (2013) Technical report, CDC—National Center for Health Statistics. https://www.cdc.gov/drugresistance/threat-report-2013/
Avinoam O, Schorb M, Beese CJ, Briggs JAG, Kaksonen M (2015) Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science (New York, NY) 348(6241):1369–1372. ISSN: 1095-9203. https://doi.org/0.1126/science.aaa9555
Curwin AJ, Brouwers N, Alonso Y Adell M, Teis D, Turacchio G, Parashuraman S, Ronchi P, Malhotra V (2016) ESCRT-III drives the final stages of CUPS maturation for unconventional protein secretion. eLife. ISSN: 2050-084X. https://doi.org/10.7554/eLife.16299
de Boer P, Hoogenboom JP, Giepmans BNG (2015) Correlated light and electron microscopy: ultrastructure lights up! Nat Methods 12(6):503–513. ISSN: 1548-7105. https://doi.org/10.1038/nmeth.3400
de Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, Lagache T, Dufour A, Olivo-Marin J-C (2012) Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 9(7):690–696. ISSN: 1548-7105. https://doi.org/10.1038/nmeth.2075
Hampoelz B, Mackmull M-T, Machado P, Ronchi P, Huy Bui K, Schieber N, Santarella-Mellwig R, Necakov A, Andrés-Pons A, Philippe JM, Lecuit T, Schwab Y, Beck M (2016) Pre-assembled nuclear pores insert into the nuclear envelope during early development. Cell 166(3):664–678. ISSN: 1097-4172. https://doi.org/10.1016/j.cell.2016.06.015
Image Registration. https://www.mathworks.com/discovery/image-registration.html
Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JAG (2011) Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol 192(1):111–119. ISSN: 1540-8140. https://doi.org/10.1083/jcb.201009037
Kukulski W, Schorb M, Kaksonen M, Briggs JAG (2012) Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell 150(3):508–520. ISSN: 1097-4172. https://doi.org/10.1016/j.cell.2012.05.046
Paul-Gilloteaux P, Heiligenstein X, Belle M, Domart M-C, Larijani B, Collinson L, Raposo G, Salamero J (2017) eC-CLEM: flexible multidimensional registration software for correlative microscopies. Nat Methods 14(2):102–103. ISSN: 1548-7105. https://doi.org/10.1038/nmeth.4170
Schellenberger P, Kaufmann R, Siebert CA, Hagen C, Wodrich H, Grünewald K (2014) High-precision correlative fluorescence and electron cryo microscopy using two independent alignment markers. Ultramicroscopy 143:41–51. ISSN: 879-2723. https://doi.org/10.1016/j.ultramic.2013.10.011
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682. ISSN: 1548-7105. https://doi.org/10.1038/nmeth.2019
Acknowledgements
We thank Marion Louveaux (Heidelberg University) for reviewing this chapter.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix: Image Transformations
Appendix: Image Transformations
6.1.1 Basic Similarity and Affine Transformations
The position of each pixel and each object inside an image is given by its two coordinates  to which an intensity value is associated.
to which an intensity value is associated.
Any linear transformation can be written as the multiplication by a matrix T that describes the transformation applied to the the image vectors.
Any linear transformation (i.e translations, rotations, scaling, etc…) can be seen as a combination of elementary transformations that can be represented as sequential matrix multiplications.
The simplest possible transformation is the uniform scaling with a constant s (◘ Fig. 6.13). The transformation can then simply be described as the Identity matrix multiplied by this constant. This means, that in order to obtain this matrix from image data, we need to find one parameter (s).
In this scaling example, if we take a scaling of s = 0.5 (i.e. reducing the image size by 2), if a pixel was at position (2,2) in the original image it would then move to position (2*0.5,2*0.5)=(1,1) in the new scaled image (see left panel of ◘ Fig. 6.13)
Another basic transformation is a rotation (◘ Fig. 6.13). Here the rotation matrix T, given a rotation angle θ, takes the form:
This means, that in order to obtain the rotation matrix from a pair of images, we need to find one parameter (θ).
Another elementary step that can happen when multi-modal images are compared is that the images are flipped with respect to each other. The transformation matrix to describe the swapping of the two coordinate axes looks like this:
Why?
Coordinates swapped!
Another elementary image transformation—the translation of coordinates—cannot be described using the aforementioned very elegant concept of nested matrix multiplications. So how can the following translation
be written as a multiplication?
In order to represent a translation as a matrix multiplication we have to add an extra dimension to our description that does not correspond to a real coordinate dimension in our image data but only plays a role during the calculations. An image coordinate would then be written like this:  . The matrix describing a translation is obtained by adding the column of the translation vector
. The matrix describing a translation is obtained by adding the column of the translation vector  to the two-dimensional Identity matrix.
to the two-dimensional Identity matrix.
The combination of rotation and translation with an optional flip of coordinate axes is called rigid transformation. A rigid transformation conserves all geometrical properties of the original structure, such as areas and relative orientations.
When adding a uniform scaling, the transformation is called similarity (◘ Fig. 6.14). Under a similarity, parallel lines remain parallel and angles are conserved. This implies that all shapes stay the same.
A general similarity can be written as:
In order to find a similarity matching two coordinate systems, the four unknown parameters s, θ, t x and t y have to be determined. The additional parameter σ which is ± 1 determines whether a coordinate flip is included or not.
When we allow a non-uniform scaling that affects the coordinate axes differently, the resulting transformation is called affine (◘ Fig. 6.15) and no longer preserves angles and shapes, but parallel lines. The representation of an affine transformation requires to define all six matrix components.
6.1.2 Higher-Order Transformations
When a coordinate frame is registered onto another coordinate system, the scaling factors that determine the stretching can be defined to vary in a linear fashion. Transformations that add this flexibility to affine transformations are called projections (◘ Fig. 6.16) and are described by a general 3 × 3 matrix with 9 unknown parameters. After a projection, straight lines will remain straight.
Instead of applying a single matrix multiplication to the coordinates, an other way of mathematically describing such coordinate transformation would be to have the result depend on higher polynomial orders of the input.
For the second order
T is a 6 × 2 matrix with 12 unknown coefficients. With higher order polynomials or groups of transformations, where each only matches a local set of coordinates, any degree of flexibility can be achieved (◘ Fig. 6.16). However, the higher the complexity of the approach, the higher the risk of generating an overfitting that only represents the priors but not the true state of the entire system.
6.1.3 Generating Transformations from Image Coordinates
The goal of a post-acquisition correlative experiment is to localize of a feature from available image data inside a second, different image dataset that provides complementary information. In order to find the transformation that registers the coordinate frame from the first imaging modality to the second, we need to define the unknown parameters. In a typical CLEM experiment, the information from the light microscopy data and those obtained by EM are fundamentally different, so an automated feature detection will most likely fail due to the lack of common structures. We therefore rely on a generic approach and on the availability of landmark pairs that the user can manually position in both image modalities.
Let’s assume we want to find the parameters of a similarity without reflection. We thus need to define the four coefficients for
with \( a=s\ast \cos \theta \) and \( b=s\ast \sin \theta \) This notation represents a set of two linear equations. Since each known coordinate pair will solve one set, we need three pairs of landmarks to solve all unknown pairs.
The higher the flexibility of the desired transformation, the higher the number of unknowns in the equations and therefore the more defined landmark pairs need to be provided.
The system of equations can thus be written as:
or in MATLAB code
1 V = T * [U;ones(1,size(U,2))]; 2 V = [x1, x2, x3; y1, y2, y3]; % x,y landmark coordinates in target frame
with the matrices U containing the source and V the target coordinates.
1 U = [x1, x2 ,x3; y1, y2, y3];
In order to solve for the matrix coefficients in T, we simply need to invert this matrix multiplication. In general, matrix multiplications are not commutative, so the order of the factors matters. In our case, we want to determine the left factor, therefore we can apply MATLAB’s “/” operator (identical to the function mrdivide).
1 T = V / [U;ones(1,size(U,2))];
However, this approach will solve the general matrix parameters and will not take into account the restrictions on our transformation matrix, such as the number of free parameters depending on the chosen type of transformation. It will simply determine a matrix that solves this set of equations. Moreover, if the number of provided landmark pairs exceeds the number of constraints necessary to solve the equations, MATLAB will find the matrix coefficients using a least-squares approach. In order to restrict the solution to a specific transformation type, we will use the MATLAB function fitgeotrans. It produces a MATLAB transformation structure that contains the matrix and some metadata. It requires the coordinate data in columns, so we transpose U and V.
1 Transformation_Type= ’nonreflectivesimilarity’; 2 structT=fitgeotrans(U’,V’,Transformation_Type); 3 T=structT.T’
The script code for these 2 methods can be found in Background_Finding Transformations.m.
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Schorb, M., Paul-Gilloteaux, P. (2020). Resolving the Process of Clathrin Mediated Endocytosis Using Correlative Light and Electron Microscopy (CLEM). In: Miura, K., Sladoje, N. (eds) Bioimage Data Analysis Workflows. Learning Materials in Biosciences. Springer, Cham. https://doi.org/10.1007/978-3-030-22386-1_6
Download citation
DOI: https://doi.org/10.1007/978-3-030-22386-1_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-22385-4
Online ISBN: 978-3-030-22386-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)