Abstract
The generic position of the enigmatic smut fungus Ustilago solida is evaluated applying molecular phylogenetic analyses using ITS and LSU rDNA sequences as well as light and scanning electron microscopical investigations of several collections of this species. Ustilago solida has previously been included in five different genera (Ustilago, Urocystis, Sorosporium, Cintractia, and Tolyposporium), however, molecular analyses revealed that this smut does not belong to any of these genera and represents a distinct ustilaginalean lineage. The closest known phylogenetic relative of Ustilago solida is Heterotolyposporium lepidospermatis, the type species of the monotypic genus Heterotolyposporium. Both smuts differ considerably in both LSU sequences and in several morphological traits, such as the structure of sori and the characteristics of spore balls. Accordingly, the new genus Shivasia is described to accommodate Ustilago solida. This smut infects different Schoenus species (Cyperaceae) in Australia and New Zealand. The description of Shivasia increases the number of endemic smut genera in Australasia to ten. Compared to all other continents the number of endemic smut genera is exceptionally high, which may point at fast evolving characters and/or may be caused by the regional history, including the long-term geographic isolation of Australasia.
Similar content being viewed by others
Introduction
The order Ustilaginales, currently classified within the subclass Ustilaginomycotina (Bauer et al. 2006), contains the largest number of smut fungi, both in terms of the number of genera and species, among all ustilaginomycotinous orders. Forty-five genera are currently included in the Ustilaginales (Begerow et al. 2007 — excluding Pseudozyma, Vánky 2008a — excluding Thecaphora; Vánky 2011, Vánky & Lutz 2011), but this number is likely to increase since at least some of these genera are undoubtedly polyphyletic (Stoll et al. 2005), and several smut species with unusual characteristics could still be wrongly classified. All members of Ustilaginales share a similar ultrastructure (Bauer et al. 1997), which accordingly is useless for generic classification. However, molecular phylogenetic analyses proved to be useful in the Ustilaginales. Indeed, molecular phylogenetic analysis has been crucial in the generic placement of several ustilaginalean species with ambiguous morphological characteristics (Piepenbring et al. 1999, Cunnington et al. 2005, Vánky et al. 2006, Bauer et al. 2007, González et al. 2007) and has also helped to elucidate the generic placement for several smuts from other orders (Vánky et al. 1998, Bauer et al. 1999, 2001a, 2005, Castlebury et al. 2005, Bauer et al. 2007, 2008, Chandra & Huff 2008, Vánky et al. 2008a, b, Lutz et al. 2012).
The generic classification of an enigmatic smut growing in the ovaries of several Schoenus species in Australasia, which had been described under the name Ustilago solida by Berkeley (in Hooker 1859), troubled several generations of smut researchers. This smut was originally described from the ovaries of “Chaetophora imberbis”, now Schoenus apogon, collected in Tasmania. Berkeley remarked that “This species connects Ustilago and Sporisporium [sic!]”. Consecutively, it was moved to Urocystis (Fischer von Waldheim 1877), Sorosporium (McAlpine 1910), and Cintractia (Piepenbring 2000), in each of these genera constituting a discordant element according to the current generic concepts (Vánky 2002). Recently, Vánky (2009) transferred Ustilago solida to Tolyposporium. Indeed, the spore balls of Ustilago solida show some resemblance to spore balls of Tolyposporium junci (type species), T. isolepidis, T. neillii, and T. piluliforme, which form a monophyletic group according to phylogenetic analyses presented by Lutz (in Vánky 2008b). This taxonomic decision could suggest that Ustilago solida reached the appropriate genus. However, since molecular data were still lacking for this species, it could not be excluded that the phenotypic similarity of spore balls may have resulted from morphological convergence, which is quite often observed in different smut fungi.
The present work aims to clarify the generic position of Ustilago solida applying molecular phylogenetic analyses using rDNA sequences as well as light and scanning electron microscopical investigation of several collections of this fungus.
Materials and Methods
Specimen sampling and documentation
The specimens examined in this study are listed in Table 1. The voucher specimens have been deposited in DAR, K, KRAM F, and H.U.V. The latter abbreviation refers to the personal collection of Kálmán Vánky, “Herbarium Ustilaginales Vánky” currently held at his home (Gabriel-Biel-Straßr 5, D-72076 Tübingen, Germany). Nomenclatural novelties were registered in MycoBank (www.MycoBank.org, Crous et al. 2004). The genetype concept follows the proposal of Chakrabarty (2010).
Morphological examination
Sorus structure, spore ball development, mature spore balls and spore characteristics were studied using dried herbarium specimens. For soral studies, young sori from herbarium specimens were rehydrated by briefly boiling in distilled water, and fixed with 2 % glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) at room temperature. Following six transfers in 0.1 M sodium cacodylate buffer, samples were postfixed in 1 % osmium tetraoxide in the same buffer for 1 h in the dark, washed in distilled water, and stained in 1 % aqueous uranyl acetate for 1 h in the dark. After five washes in distilled water, samples were dehydrated in acetone, using 10 min changes at 25 %, 50 %, 70 %, 95 %, and three times in 100 % acetone. Samples were embedded in Spurr’s plastic and sectioned with a diamond knife. Semi-thin sections were transferred to a microscope slide, stained with new fuchsin and crystal violet, mounted in Entellan under a cover slip, and studied by light microscopy (LM) at various magnifications.
For LM, spore balls and spores were dispersed in a droplet of lactophenol on a microscope slide, covered with a cover slip, gently heated to boiling point to rehydrate the spores and to eliminate air bubbles, and examined at 1000 × magnification. For examination of spore ball development, sori were boiled in a mixture of lactophenol with cotton blue and distilled water, and hand sectioned with a razor blade under a stereomicroscope. Pieces of host tissues from the basal part of the sori and very young spore balls were transferred into a droplet of lactophenol with cotton blue and covered with a cover slip. Gentle pressure was applied until the host tissue became flat. Air bubbles were eliminated by gently heating to boiling point.
For scanning electron microscopy (SEM), spore balls and spores were mounted on carbon tabs and fixed to an aluminium stub with double-sided transparent tape. The stubs were sputter-coated with carbon using a Cressington sputtercoater and viewed under a Hitachi S-4700 scanning electron microscope, with a working distance of ca. 11 mm. SEM micrographs were taken in the Laboratory of Field Emission Scanning Electron Microscopy and Microanalysis at the Institute of Geological Sciences of Jagiellonian University, Krakow (Poland).
DNA extraction, PCR, and sequencing
Genomic DNA was isolated directly from the herbarium specimens. For methods of isolation and crushing of fungal material, DNA extraction, amplification, purification of PCR products, sequencing, and processing of the raw data see Lutz et al. (2004). ITS 1 and ITS 2 regions of the rDNA including the 5.8S rDNA (ITS, about 780 bp) were amplified using the primer pair M-ITS1 (Stoll et al. 2003) and ITS4 (White et al. 1990). The 5′-end of the nuclear large subunit ribosomal DNA (LSU, about 650 bp) was amplified using the primer pair NL1 and NL4 (O’Donnell 1993). PCR primers were also used for cycle sequencing. For amplification the annealing temperature was adjusted to 45 °C. DNA sequences determined for this study were deposited in GenBank. GenBank accession numbers are given in Table 1 and Fig. 1.
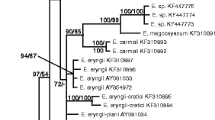
Bayesian inference of phylogenetic relationships within the sampled Ustilaginomycetes: Markov chain Monte Carlo analysis of an alignment of LSU sequences using the GTR+I+G model of DNA substitution with gamma distributed substitution rates and an estimated proportion of invariant sites, random starting trees and default starting parameters of the DNA substitution model. A 50 % majority-rule consensus tree is shown computed from 45 000 trees that were sampled after the process had reached stationarity. The topology was rooted with the exobasidiomycetous species Entyloma microsporum, Exobasidium vaccinii, and Tilletia caries. Numbers on branches before slashes are estimates for a posteriori probabilities, numbers on branches after slashes are ML bootstrap support values. Branch lengths were averaged over the sampled trees. They are scaled in terms of expected numbers of nucleotide substitutions per site. The taxonomic concept used here follows Bauer et al. (2001b).
Phylogenetic analyses
The Ustilago solida specimens examined in this study are listed in Table 1. For molecular phylogenetic analyses the following sequences from GenBank were additionally used (Begerow et al. 1997, Piepenbring et al. 1999, Castlebury et al. 2005, Hendrichs et al. 2005, Stoll et al. 2005, Matheny et al. 2006, Vánky et al. 2006, Bauer et al. 2007, Begerow et al. 2007, González et al. 2007, Vánky & Lutz 2007, Bauer et al. 2008, Vánky & Lutz, in Vánky 2008b, Vánky et al. 2008b, Kottke et al. 2010, Lutz et al. 2012): Anomalomyces panici DQ459347, Antherospora vaillantii EF653980, Anthracoidea caricis AY563589, Cintractia axicola DQ631906, Dermatosorus cyperi AJ236157, Doassansiopsis deformans AF009849, Entyloma microsporum DQ185435, Eriomoeszia eriocauli AY740094, Exobasidium vaccinii FJ644526, Farysia chardoniana AF009859, Flamingomyces ruppiae DQ185436, Floromyces anemarrhenae EU221284, Franzpetrakia microstegii GU139170, Heterotolyposporium lepidospermatis DQ875362, Leucocintractia scleriae AJ236154, Macalpinomyces eriachnes AY740090, Melanopsichium pennsylvanicum AY740093, Melanotaenium endogenum DQ789979, Melanoxa oxalidis EF635908, Melanustilospora ari EF517924, Moesziomyces bullatus DQ875365, Moreaua fímbristylidis DQ875367, Mundkurella kalopanacis AF009869, Mycosyrinx cissi DQ875368, Parvulago marina DQ185437, Pericladium grewiae DQ875370, Portalia uljanishcheviana EF118824, Restiosporium meneyae DQ875371, Schizonella melanogramma AF009870, Sporisorium sorghi AF009872, Stegocintractia luzulae AJ236148, Thecaphora saponariae EF200047, T. seminis-convolvuli AF009874, Tilletia caries AY819007, Tolyposporium isolepidis EU246949, T. junci AF009876, T. neillii EU246952, T. piluliforme AF009871, Tranzscheliella hypodytes DQ875373, Trichocintractia utriculicola AF009877, Urocystis ficariae EF517939, Ustacystis waldsteiniae AF009880, Ustanciosporium standleyanum DQ846888, Ustilago hordeiAY740122, Vankya ornithogali EF517926, and Websdanea lyginiae AJ236159.
To elucidate the phylogenetic position of the Ustilago solida specimens their LSU sequences were analysed within a dataset covering all urocystidalean and ustilaginalean genera of which sequences were available in GenBank. If present in GenBank the respective type species were used. Additionally, all Tolyposporium LSU sequences available in GenBank and the LSU sequence of Thecaphora saponariae (type species of Sorosporium) were added.
Sequence alignment was obtained using MAFFT 6.853 (Katoh et al. 2002, 2005, Katoh & Toh 2008) using the L-INS-i option. To obtain reproducible results, manipulation of the alignment by hand as well as manual exclusion of ambiguous sites were avoided as suggested by Giribet & Wheeler (1999) and Gatesy et al. (1993), respectively. Instead, highly divergent portions of the alignment were omitted using GBlocks 0.91b (Castresana 2000) with the following options. ‘Minimum Number of Sequences for a Conserved Position’: 25, ‘Minimum Number of Sequences for a Flank Position’: 25, ‘Maximum Number of Contiguous Non-conserved Positions’: 8, ‘Minimum Length of a Block’: 5, and ‘Allowed Gap Positions’ to ‘With half’.
The resulting alignment (new number of positions: 586; 21 % of the original 2790 positions; numberof variable sites: 312) was used for phylogenetic analyses using a Bayesian Approach and Maximum Likelihood (ML). A Bayesian approach using a Markov chain Monte Carlo (MCMC) technique was used as implemented in the computer program MrBayes 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003). Four incrementally heated simultaneous Markov chains were run over 5 M generations using the general time reversible model of DNA substitution with gamma distributed substitution rates and an estimated proportion of invariant sites, random starting trees and default starting parameters of the DNA substitution model as recommended by Huelsenbeck & Rannala (2004). Trees were sampled every 100th generation resulting in an overall sampling of 50 001 trees. From these, the first 5 001 trees were discarded (burnin = 5 001). The trees sampled after the process had reached stationarity (45 000 trees) were used to compute a 50 % majority rule consensus tree to obtain estimates for the a posteriori probabilities of groups of species. This Bayesian approach of phylogenetic analysis was repeated five times to test the independence of the results from topological priors (Huelsenbeck et al. 2002). ML analysis (Felsenstein 1981) was conducted with the RAxML 7.2.8 software (Stamatakis 2006), using raxmlGUI (Silvestro & Michalak 2012), invoking the GTRCAT and the rapid bootstrap option (Stamatakis et al. 2008) with 1000 replicates.
Trees were rooted with the exobasidiomycetous species Entyloma microsporum, Exobasidium vaccinii, and Tilletia caries.
Results
Morphology
The morphological characteristics of Ustilago solida are included in the species description and depicted in illustrations (Figs 2–4).
Shivasia solida on Schoenus apogon. A. Embedded, stained, semi-thin section of a sorus (H.U.V. 17649), sh = sporogenous hyphae, sp = spore balls, p = peridium. B. Fungal cells of the peridium covering the sori, formed of thick-walled, sterile hyphae (H.U.V. 15059). C. Spore ball formation in sporogenous fungal layer on the surface of innermost floral organs, in U-shaped pockets, hand sectioned, stained with cotton blue in lactophenol (H.U.V. 15059). D–E. Young spores and spore balls covered by fungal cells of the young peridium, embedded in plastic, sectioned and stained with new fuchsin and cristal violet (H.U.V. 15059). F. Spore balls in different developmental stages, hand sectioned, stained with cotton blue in lactophenol (H.U.V. 15059). G–H. Spore germination in water, at room temperature, in 3–5 days (H.U.V. 15059). Bars: A = 100 μm, B–H = 10 μm.
Shivasia solida on Schoenus apogon (KRAM F-49115). A–C. Spore balls and spores seen in LM, note mucilaginous layer (subhyaline caps) around spores marked by arrows. D–E. Spore balls and spores seen in SEM, note that spores are enclosed by remnants of mucilaginous layer that form (pseudo-)ornamentation, while the spore surface is verruculose as marked by arrow. Bars = 10 μm.
Phylogenetic analyses
The LSU sequences of the two Ustilago solida specimens analysed were identical. Compared to the LSU of Heterotolyposporium lepidospermatis DQ875362 they differed in 51 positions (9.75 %) in 36 different sections.
BLAST searches (Altschul et al. 1997) for the ITS sequence of Ustilago solida H.U.V. 17649 revealed closest similarity to Tolyposporium isolepidis EU246950 and T. neillii EU246951. However, ITS sampling of ustilaginaceous species in GenBank is limited (e.g., there is no Heterotolyposporium lepidospermatis ITS sequence available) and sequences differ to a great extent (e.g., compared to the ITS of Tolyposporium isolepidis and T. neillii the ITS sequence of Ustilago solida differed in 211 positions (27.51 %) in 123 different sections). Thus, molecular phylogenetic analyses of the available ITS sequences, similar to the LSU analyses, supported the ustilaginaceous affiliation of Ustilago solida but did not resolve the placement within the Ustilaginaceae (data not shown).
The different runs of the Bayesian phylogenetic analyses that were performed and the ML analyses yielded consistent topologies. To illustrate the results, the consensus tree of one run of the Bayesian phylogenetic analyses is presented (Fig. 1). Estimates for a posteriori probabilities are indicated on branches before slashes, numbers on branches after slashes are ML bootstrap support values.
In all analyses the two Ustilago solida specimens formed a cluster within the Ustilaginaceae sensu Bauer et al. (2001b), which was revealed in a group consisting of Heterotolyposporium lepidospermatis, and the cluster of Farysia chardoniana, Schizonella melanogramma, and Stegocintractia luzulae. A closer relation of the Ustilago solida specimens to any of the genera Cintractia, Sorosporium (here: Thecaphora saponariae, the type species of Sorosporium), Tolyposporium, Urocystis or Ustilago was not revealed by any of the phylogenetic analyses.
Taxonomy
Molecular phylogenetic analyses, sorus, spore ball and spore morphology, as well as the type of spore germination, supplemented by host plant taxonomy give evidence that a new genus should be erected to accommodate Ustilago solida.
Shivasia Vánky, M. Lutz & M. Piątek, gen. nov. MycoBank MB800821
Etymology: This genus is named in the honour of Roger Graham Shivas, a multi-talented Australian mycologist, interested in plant parasitic microfungi, author of two books and over 130 scientific papers, in which over 160 new species and several new genera are described. Roger is a sharp-eyed, ardent collector who collected not only in Australia but also in Bolivia, Burma, China, India, Indonesia, Malaysia, New Zealand, Norway, Papua New Guinea, Philippines, South Africa, Thailand, and Vietnam.
Type species: Shivasia solida (Berk.) Vánky, M. Lutz & M. Piątek 2012. Sori in the flowers of Schoenus (Cyperaceae) forming hard, globoid, black bodies composed of spore balls and spores on the surface of innermost floral organs developed in sporogenous hyphae within U-shaped pockets, at first covered by a fungal peridium. Spore balls few to many-spored, composed of spores only, enclosed by a subhyaline mucilaginous layer. Spores pigmented (brown). Sterile cells absent.
Shivasia solida (Berk.) Vánky, M. Lutz & M. Piątek, comb. nov.
MycoBank MB800822
-
Basionym: Ustilago solida Berk., in Hooker, Flora Tasmaniae 2: 270 (1859).
-
Synonyms: Urocystis solida (Berk.) A.A. Fisch. Waldh., Aperçu Syst. Ustil.: 38 (1877).
-
Sorosporium solidum (Berk.) McAlpine, Smuts of Australia: 185 (1910).
-
Cintractia solida (Berk.) M. Piepenbr., Nova Hedwigia 70: 310 (2000).
-
Tolyposporium solidum (Berk.) Vánky, Mycotaxon 110: 320 (2009).
Type: [Australia: Tasmania]: “Herb. Berk. 4751, 1879. Ustilago solida, Berk. on Chaetospora imberbis Penquite 21/12/45 h°581.” [original label of holotype specimen, Fig. 2a] (K(M) 171338 — holotype; DAR 59818 — microscope slide ex-holotype). — Australia: Tasmania: 170 km NE of Hobart, on Schoenus apogon, 8 March 1996, C. Vánky & K. Vánky (H.U.V. 17649 — epitype designated here; TUB 20001 — isoepitype).
Sori (Figs 2–3) in all flowers of an inflorescence, comprising the innermost floral organs, visible between the glumes as black, globose to ovoid bodies, 1–2 mm diam, rarely also on the stems, then fusiform, at first covered by a thick, whitish brown fungal peridium of thick-walled, sterile hyphae that early flakes away exposing the compact mass of spore balls with spores, powdery on the surface. Spore balls (Fig. 4) usually irregular or globoid to ellipsoidal, composed of 2–15 spores, loose but rather permanent, 25–55(–70) × 20–10 μm, reddish brown, enclosed by subhyaline mucilaginous layer. Spores (Fig. 4) subglobose, ovoid, elongate or irregular, flattened on one or two sides, 15–20 × 12–16 μm, yellowish to pale reddish brown; wall uneven, 0.5–1.5 μm thick, smooth to rough, in SEM finely, densely, irregularly verruculose and covered by remnants of the mucilaginous layer which form irregularly warty (pseudo-)ornamentation. Spore balls and spores produced on the surface of host tissues in hyaline, sporogenous fungal layer within radially arranged, U-shaped pockets (Fig. 3C–F). Spore germination (Fig. 3G–H; on water, at room temperature, in 3–5 d) results in long, aseptate basidia on which apically elongated, cylindrical basidiospores are produced that germinate by ilaments.
The ITS/LSU epigenetype sequences are deposited in GenBank with the accession numbers JF966731/JF966730, respectively.
Hosts: On different Schoenus species (Cyperaceae): S. apogon, S. calyptratus, S. carsei, S. cruentus, S. latelaminatus, S. maschalinus, S. nanus, S. nitens var. concinnus, S. pauciflorus, S. tesquorum, and Schoenus sp. (Table 1, Vánky & McKenzie 2002, Vánky & Shivas 2008).
Distribution: The genus and species are restricted to southeastern Australasia: south-east Australia, including Tasmania, and north-west New Zealand (Fig. 5, based on the specimens included in Table 1 as well as in Vánky & McKenzie 2002, and Vánky & Shivas 2008).
Discussion
In the present study molecular phylogenetic analyses and morphological data were used to clarify the systematic position of Ustilago solida. The molecular analyses revealed that this smut does not belong to any genus in which it was included during the last 150 years. The morphological characteristics also contradict the placement of this species in Ustilago, Urocystis, Sorosporium as well as in Cintractia.
In the past the generic concept of Ustilago was very broad (Zundel 1953), but now this genus is restricted to smuts infecting poacean hosts, producing sori in vegetative or generative host plant organs, and having sori composed only of single spores, never forming spore balls (Vánky 2002). The species included in the genus Urocystis infect a wide range of host species from both mono- and dicotyledonous families, and are characterised by the production of spores in spore balls, which are usually enclosed by a complete or incomplete layer of sterile cells (Vánky 2002). The genus Sorosporium included in the past many, often unrelated smuts having spore balls (Zundel 1953), but the generic type, Sorosporium saponariae, has features of the genus Thecaphora with which it was merged based on both morphological and molecular phylogenetic analyses (Vánky 1998a, b, Vánky & Lutz 2007, Vánky et al. 2008a). Thecaphora (incl. Sorosporium) is restricted to smut fungi parasitic on dicotyledonous host plants, having sori with a granular-powdery spore ball mass that is yellowish, pale brown or dark reddish-brown, but never black or almost black (Vánky 2002, Vánky et al. 2008a). Cintractia species infect cyperaceous host plants forming single spores only (Vánky 2002, Piqtek & Vánky 2007). The placement of Ustilago solida in Cintractia was in any case considered as provisional (Piepenbring 2000).
The most surprising result of the molecular analyses is that Ustilago solida is also not a member of the genus Tolyposporium, with which it shares many morphological traits, including the formation of sori in the floral organs and stems, sporulation within U-shaped pockets formed on the sterile stroma, and the development of spore balls (Piepenbring 2000, Vánky 2002). However, there are morphological differences between Ustilago solida and Tolyposporium that support the affiliation to different phylogenetic lineages. Ustilago solida differs from the type species of Tolyposporium (i.e. T. junci) in spore wall ornamentation (rough or finely verruculose vs. irregularly, coarsely warty) and most notably in that spore balls of U. solida are enclosed by a mucilaginous layer that in light microscope is seen as subhyaline cap on the outer sides of spores, and in scanning electron micrographs as an irregularly warty (pseudo-) ornamentation. A mucilaginous layer around the spore balls is absent in Tolyposporium. This is in line with the observation of Piepenbring (2000) who used these two characteristics to differentiate Tolyposporium from Cintractia solida. It is worth noting that these two diagnostic features of Tolyposporium are easily applied to that three remaining species currently classified in this genus (T. isolepidis, T. neillii, and T. piluliforme).
The molecular analyses revealed that the closest phylogenetic relative of Ustilago solida may be Heterotolyposporium lepidospermatis, the type of the monotypic genus Heterotolyposporium. However, these two smuts differ considerably in LSU base sequences as well as in several morphological characteristics. In particular, the structure of the sori could hardly be considered congeneric. Heterotolyposporium lepidospermatis develops sori in the inflorescence of Lepidosperma ensiforme, with powdery spore masses replacing all central floral organs. A peridium is lacking and the sori are covered only by the outermost 3–4 glumes (Vánky 1997, 2002). This contrasts with the sori of Ustilago solida that are coated, at least in young stages, by a peridium, and the innermost floral organs are covered by a sterile stroma with U-shaped pockets within which spore balls are produced. The characteristics of the spore balls that differentiate Ustilago solida and Tolyposporium also distinguish Ustilago solida and Heterotolyposporium. The presence of two kinds of spores, originally used as the main diagnostic feature of Heterotolyposporium (Vánky 1997), is systematically irrelevant at the genus level since this character evolved convergently in Tolyposporium piluliforme, which is only distantly related to Heterotolyposporium (Piepenbring et al. 1999, Vánky & Lutz, in Vánky 2008b).
Both genetic and morphological features reveal Ustilago solida as a unique smut that should be accommodated in a distinct genus, described here as Shivasia. This is in line with the current generic concept in smut fungi where distinct phylogenetic lineages distinguishable by morphological and/ or ecological characters are referred to distinct genera (e.g., Piepenbring et al. 1999, Vánky et al. 2006, 2008b, Lutz et al. 2012). The close phylogenetic relation of Heterotolyposporium and Shivasia is reflected in the close phylogenetic relation of their host genera. Both Lepidosperma and Schoenus are placed in the same tribe Schoeneae within the Cyperaceae (Verboom 2006, Simpson et al. 2007, Muasya et al. 2009). Thus co-speciation of parasites and hosts may have played a role in the evolution of these two smuts.
In addition to resolving the phylogenetic and systematic placement of Ustilago solida, this study reports an emerging number of smut genera currently known exclusively from Australasia. Based on the present knowledge, and considering the relatively uniform and stabilized generic concept in smut fungi, they may be treated as endemic to this ecozone. The high rate of endemic smuts in Australia has been discussed at length by Shivas & Vánky (2003) and less extensively also by Vánky & Shivas (2008). Amongst the 309 species reported from the continent (Vánky & Shivas 2008, Barrett et al. 2009, McTaggart & Shivas 2009a, b, Shivas & McTaggart 2009, Vánky 2009, Shivas et al. 2010, Piqtek & Shivas 2011, Shivas et al. 2011, Crous et al. 2012) about half are endemic. Six genera were considered to be endemic to Australia by Vánky & Shivas (2008), namely Anomalomyces, Centrolepidosporium, Farysporium, Fulvisporium, Pseudotracya, and Websdanea, but in fact Farysporium should be deleted as a strictly Australian endemic since it also occurs in New Zealand. This removal is balanced by adding Heterotolyposporium because, in the present circumscription, that genus has only one species, H. lepidospermatis, known from only one locality in Tasmania. Four additional genera are endemic to Australasia, being present in both Australia and New Zealand (Farysporium, Restiosporium, and Shivasia) or in Australia and Papua New Guinea (Macalpinomyces). Until recently Macalpinomyces included many unrelated smuts from different parts of the globe, but recent molecular analyses (Stoll et al. 2005) narrowed this genus to the type species M. eriachnes that occurs exclusively in Australasia. Thus, ten endemic genera occur in the Australasian ecozone and this number is not comparable with any other continent since all of them have lower numbers of endemic smut genera (Table 2). The high number of unique smut genera may point to rapidly evolving characters and/or may result from the regional history, including the long geographic isolation of Australasia which started with the break-up of Gondwana and the initial separation of Australia and Tasmantia (incl. New Zealand) terranes from Antarctic terranes about 96 Myr and 84 Myr ago, respectively (McLoughlin 2001).
Shivasia solida has been reported on ten different Schoenus species. Recent molecular analyses of different smut genera (Lutz et al. 2005, Carris et al. 2007, Vánky & Lutz 2007, Bauer et al. 2008, Lutz et al. 2008, Kemler et al. 2009, Piatek et al. 2011, Lutz et al. 2012, Piqtek et al. 2012, Savchenko et al. 2012) revealed that such polyphagous species are usually complexes of morphologically similar cryptic species that are often restricted to single host plant species. However, in the present study sequences were obtained only from specimens on Schoenus apogon, while repeated attempts to obtain sequences from specimens on two other hosts (Schoenus pauciflorus, H.U.V. 16755, and Schoenus tesquorum, H.U.V. 20073) failed. Thus, the question whether the collections on other Schoenus species than Schoenus apogon are genetically identical or cryptic species exist within what we now consider as one species is left open for future studies. To stabilise the taxonomy of Shivasia solida and to make future genetic comparisons possible an epitype is selected here based on the sequenced specimen collected on the same host (Schoenus apogon) in the same geographical area (Tasmania) as the holotype. This is in concordance with recent recommendations for epitypifications (Hyde & Zhang 2008).
The genus Schoenus shows greatest species diversity in Australasia. This is reflected by the inhabiting smut fungi — amongst seven smuts known on this genus worldwide, five occur in Australasia (Vánky & Websdane 1995, Vánky 2009). The two extralimital species are Anthracoidea andina in southern South America (Argentina: Tierra del Fuego) and Moreaua kochiana in Central Europe (Italy and Switzerland). The Schoenus smuts belong to three genera, Anthracoidea, Moreaua and Shivasia, that are not closely related, suggesting that at least three independent colonisation events took place onto this host plant genus in the course of evolution.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402
Barrett MD, Barrett RL, Shivas RG, McTaggart AR (2009) Fungal Planet no. 33. Tilletia micrairae. Persoonia 22, 170–171
Bauer R, Begerow D, Nagler A, Oberwinkler F (2001a) The Georgefischeriales: a phylogenetic hypothesis. Mycological Research 105, 416–424
Bauer R, Begerow D, Oberwinkler F, Piepenbring M, Berbee ML (2001b) Ustilaginomycetes. In: The Mycota. Vol. VII, Part B. Systematics and Evolution (DJ McLaughlin, EG McLaughlin, PA Lemke, eds): 57–83. Berlin: Springer-Verlag.
Bauer R, Begerow D, Sampaio JP, Oberwinkler F (2006) The simple-septate basidiomycetes. Mycological Progress 5, 41–66
Bauer R, Begerow D, Vánky K, Oberwinkler F (1999) Ustilaginomycetes on Selaginella. Mycologia 91, 475–484
Bauer R, Lutz M, Begerow D, Piqtek M, Vénky K, Bacigalova K, Oberwinkler F (2008) Anther smut fungi on monocots. Mycological Research 112, 1297–1306
Bauer R, Lutz M, Oberwinkler F (2005) Gjaerumia, a new genus in the Georgefischeriales (Ustilaginomycetes). Mycological Research 109, 1250–1258
Bauer R, Lutz M, Piątek M, Vánky K, Oberwinkler F (2007) Flamingomyces and Parvulago, new genera of marine smut fungi (Ustilaginomycotina). Mycological Research 111, 1199–1206
Bauer R, Oberwinkler F, Vánky K (1997) Ultrastructural markers and systematics in smut fungi and allied taxa. Canadian Journal of Botany 75, 1237–1314
Begerow D, Bauer R, Oberwinkler F (1997) Phylogenetic studies on nuclear LSU rDNA sequences of smut fungi and related taxa. Canadian Journal of Botany 75, 2045–2056
Begerow D, Stoll M, Bauer R (2007(‘2006’). A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98, 906–916
Carris LM, Castlebury LA, Huang GM, Alderman SC, Luo JF, Bao XD (2007) Tilletia vankyi, a new species of reticulate-spored bunt fungus with non-conjugating basidiospores infecting species of Festuca and Lolium. Mycological Research 111, 1386–1398
Castlebury LA, Carris LM, Vánky K (2005) Phylogenetic analysis of Tilletia and allied genera in order Tilletiales (Ustilaginomycetes; Exobasidiomycetidae) based on large subunit nuclear rDNA sequences. Mycologia 97, 888–900
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17, 540–552
Chakrabarty P (2010) Genetypes: a concept to help integrate molecular phylogenetics and taxonomy. Zootaxa 2632, 67–68
Chandra A, Huff DR (2008) Salmacisia, a new genus of Tilletiales: reclassification of Tilletia buchloëana causing induced hermaphroditism in buffalograss. Mycologia 100, 81–93
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50, 19–22
Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GESJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ (2012) Fungal Planet description sheets: 107–127. Persoonia 28, 138–182
Cunnington JH, Vánky K, Shivas RG (2005) Lundquistia is a synonym of Sporisorium (Ustilaginomycetes). Mycologia Balcanica 2, 95–99
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution 17, 368–376
Fischer von Waldheim AA (1877) Aperçu systématique des Ustilaginées, leurs plantes nourricières et la localisation de leurs spores. Paris: Lahure.
Gatesy J, DeSalle R, Wheeler W (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Molecular Phylogenetics and Evolution 2, 152–157
Giribet G, Wheeler WC (1999) On gaps. Molecular Phylogenetics and Evolution 13, 132–143
González V, Vánky K, Platas G, Lutz M (2007) Portalia gen. nov. (Ustilaginomycotina). Fungal Diversity 27, 45–59
Hendrichs M, Begerow D, Bauer R, Oberwinkler F (2005) The genus Anthracoidea (Basidiomycota, Ustilaginales): a molecular phylogenetic approach using LSU rDNA sequences. Mycological Research 109, 31–40
Hooker JD (1858–59) The botany of the Antarctic Voyage. Vol. 3. Flora Tasmaniae. Vol. 2. London: Reeve.
Huelsenbeck JP, Larget B, Miller RE, Ronquist F (2002) Potential applications and pitfalls of Bayesian inference of phylogeny. Systematic Biology 51, 673–688
Huelsenbeck JP, Rannala B (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Systematic Biology 53, 904–913
Huelsenbeck JP, Ronquist F (2001) MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics Applications Note 17, 754–755
Hyde KD, Zhang Y (2008) Epitypification: should we epitypify? Journal of Zhejiang University Science B 9, 842–846
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33, 511–518
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30, 3059–3066
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program (outlines version 6). Briefings in Bioinformatics 9, 286–298
Kemler M, Lutz M, Göker M, Oberwinkler F, Begerow D (2009) Hidden diversity in the non-caryophyllaceous plant-parasitic members of Microbotryum (Pucciniomycotina: Microbotryales). Systematics and Biodiversity 7, 297–306
Kottke I, Suarez JP, Herrera P, Cruz D, Bauer R, Haug I, Garnica S (2010) Atractiellomycetes belonging to the ‘rust’ lineage (Pucciniomycotina) form mycorrhizae with terrestrial and epiphytic neotropical orchids. Proceedings of the Royal Society of London, B: Biological Sciences 277, 1289–1298
Lutz M, Bauer R, Begerow D, Oberwinkler F, Triebel D (2004) Tuberculina, rust relatives attack rusts. Mycologia 96, 614–626
Lutz M, Göker M, Piatek M, Kemler M, Begerow D, Oberwinkler F (2005) Anther smuts of Caryophyllaceae: molecular characters indicate host-dependent species delimitation. Mycological Progress 4, 225–238
Lutz M, Piątek M, Kemler M, Chlebicki A, Oberwinkler F (2008) Anther smuts of Caryophyllaceae: molecular analyses reveal further new species. Mycological Research 112, 1280–1296
Lutz M, Vánky K, Bauer R (2012) Melanoxa, a new genus in the Urocystidales (Ustilaginomycotina). Mycological Progress 11, 149–158
Matheny PB, Gossmann JA, Zalar P, Arun Kumar TK, Hibbett DS (2006) Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Canadian Journal of Botany 84, 1794–1805
McAlpine D (1910) The Smuts of Australia: their structure, life history, treatment, and classification. Melbourne: J. Kemp.
McLoughlin S (2001) The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany 49, 271–300
McTaggart AR, Shivas RG (2009a) Fungal Planet No. 36. Tilletia challinorae. Persoonia 23, 184–185
McTaggart AR, Shivas RG (2009b) Fungal Planet No. 37. Macalpinomyces mackinlayi. Persoonia 23, 186–187
Muasya AM, Simpson DA, Verboom GA, Goetghebeur P, Naczi RFC, Chase MW, Smets E (2009) Phylogeny of Cyperaceae based on DNA sequence data: current progress and future prospects. Botanical Review 75, 2–21
O´Donnell KL (1993) Fusarium and its near relatives. In: The Fungal Holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics (DR Reynolds, JW Taylor, eds): 225–233. Wallingford: CAB International.
Piatek M, Lutz M, Ronikier A, Kemler M, Swiderska-Burek U (2012) Microbotryum heliospermae, a new anther smut fungus parasitic on Heliosperma pusillum in the mountains of the European Alpine System. Fungal Biology 116, 185–195
Piatek M, Lutz M, Smith PA, Chater AO (2011) A new species of Antherospora supports the systematic placement of its host plant. IMA Fungus 2, 135–142
Piatek M, Shivas RG (2011) Sporisorium warambiense sp. nov, a fourth smut fungus on Xerochloa in Australia. Mycological Progress 10, 57–60
Piatek M, Vánky K (2007) Cintractia bulbostylidicola sp. nov. (Ustilaginomycotina) from North America. Nova Hedwigia 85, 187–194
Piepenbring M (2000) The species of Cintractia s. l. (Ustilaginales, Basidiomycota). Nova Hedwigia 70, 289–372
Piepenbring M, Begerow D, Oberwinkler F (1999) Molecular sequence data assess the value of morphological characteristics for a phylogenetic classification of species of Cintractia. Mycologia 91, 485–498
Ronquist FR, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574
Savchenko KG, Lutz M, Piqtek M, Heluta VP, Nevo E (2012) Anthracoidea caricis-meadii is a new North American smut fungus on Carex sect. Paniceae. Mycologia: DOI: 10.3852/12-074
Shivas RG, Barrett MD, Barrett RL, Vánky K (2011) Two new species of Moreaua (Ustilaginomycetes), on Actinoschoenus and Chrysitrix, from Western Australia. Mycologia Balcanica 8, 137–140
Shivas RG, McTaggart AR, Ryley MJ, Scharaschkin T, Gambley CF (2010) First report of the smut fungus Ustanciosporium appendiculatum in Australia. Australasian Plant Disease Notes 5, 17–18
Shivas RG, McTaggart AR (2009) Three new species of Tilletia on native grasses from northern Australia. Australasian Plant Pathology 38, 128–131
Shivas RG, Vánky K (2003) Biodiversity of Australian smut fungi. Fungal Diversity 13, 137–152
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution 12, 335–337
Simpson DA, Muasya AM, Alves MV, Bruhl JJ, Dhooge S, Chase MW, Furness CA, Ghamkhar K, Goetghebeur P, Hodkinson TR, Marchant AD, Reznicek AA, Nieuwborg R, Roalson EH, Smets E, Starr JR, Thomas WW, Wilson KL, Zhang XF (2007) Phylogeny of Cyperaceae based on DNA sequence data — a new rbcL analysis. Aliso 23, 72–83
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57, 758–771
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690
Stoll M, Begerow D, Oberwinkler F (2005) Molecular phylogeny of Ustilago, Sporisorium, and related taxa based on combined analyses of rDNA sequences. Mycological Research 109, 342–356
Stoll M, Piepenbring M, Begerow D, Oberwinkler F (2003) Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales), based on internal transcribed spacer (ITS) sequences. Canadian Journal of Botany 81, 976–984
Vánky K (1997) Heterotolyposporium, a new genus of Ustilaginales. Mycotaxon 63, 143–154
Vánky K (1998a) Proposal to conserve the generic name Thecaphora Fingerhuth against Sorosporium Rudolphi (Fungi, Ustilaginales). Taxon 47, 153–154
Vánky K (1998b) Taxonomical studies on Ustilaginales — XVIII. Mycotaxon 69, 93–116
Vánky K (2002) Illustrated Genera of Smut Fungi, 2nd edn. St Paul, MN: American Phytopathological Society Press.
Vánky K (2008a) Smut fungi (Basidiomycota p.p., Ascomycota p.p.) of the world: novelties, selected examples, trends. Acta Microbiologica et Immunologica Hungarica 55, 91–109
Vánky K (2008b) Taxonomic studies on Ustilaginomycetes — 28. Mycotaxon 106, 133–178
Vánky K (2009) Taxonomic studies on Ustilaginomycetes — 29. Mycotaxon 110, 289–324
Vánky K (2011) Bambusiomyces, a new genus of smut fungi (Ustilaginomycetes). Mycologia Balcanica 8, 141–145
Vánky K, Bauer R, Begerow D (1998) Doassinga, a new genus of Doassansiales. Mycologia 90, 964–970
Vánky K, Lutz M, Bauer R (2008a) About the genus Thecaphora (Glomosporiaceae) and its new synonyms. Mycological Progress 7, 31–39
Vánky K, Lutz M, Bauer R (2008b) Floromyces, a new genus of Ustilaginomycotina. Mycotaxon 104, 171–184
Vánky K, Lutz M, Shivas RG (2006) Anomalomyces panici, new genus and species of Ustilaginomycetes from Australia. Mycologia Balcanica 3, 119–126
Vánky K, Lutz M (2007) Revision of some Thecaphora species (Ustilaginomycotina) on Caryophyllaceae. Mycological Research 111, 1207–1219
Vánky K, Lutz M (2011) Tubisorus, a new genus of smut fungi (Ustilaginomycetes) for Sorosporium pachycarpum. Mycologia Balcanica 8, 129–135
Vánky K, McKenzie EHC (2002) Smut Fungi of New Zealand. [Fungi of New Zealand. Vol. 2.]: Fungal Diversity Press.
Vánky K, Shivas RG (2008) Fungi of Australia: the smut fungi. Melbourne: CSIRO Publishing.
Vánky K, Websdane K (1995) Ustilaginales of Schoenus (Cyperaceae). Mycotaxon 56, 217–229
Verboom GA (2006) A phylogeny of the schoenoid sedges (Cyperaceae: Schoeneae) based on plastid DNA sequences, with special reference to the genera found in Africa. Molecular Phylogenetics and Evolution 38, 79–89
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (MA Innis, DH Gelfand, JJ Sninsky, TJ White, eds): 315–322. San Diego: Academic Press.
Zundel GL (1953) The Ustilaginales of the world. Pennsylvania State College School of Agriculture Department of Botany Contribution 176: xi+1–410.
Acknowledgements
We thank Michael Weiß, Sigisfredo Garnica, and Robert Bauer (Tübingen) for providing facilities for molecular analyses, Michael
Weiß for critically reading the manuscript, Christine Vánky (Tübingen) fortechnical assistance with the illustrations, Jolanta Piatek (Krakow) for preparing the world map, Magda Wagner-Eha (Tübingen) for technical assistance with the semi-thin sections, Anna tatkiewicz (Krakow) for help with the SEM photomirographs, and the Curators of DAR and K (M) for the loan of specimens.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lutz, M., Vánky, K. & Piątek, M. Shivasia gen. nov. for the Australasian smut Ustilago solida that historically shifted through five different genera. IMA Fungus 3, 143–154 (2012). https://doi.org/10.5598/imafungus.2012.03.02.06
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.5598/imafungus.2012.03.02.06