Abstract
Geodermatophilus obscurus Luedemann 1968 is the type species of the genus, which is the type genus of the family Geodermatophilaceae. G. obscurus is of interest as it has frequently been isolated from stressful environments such as rock varnish in deserts, and as it exhibits interesting phenotypes such as lytic capability of yeast cell walls, UV-C resistance, strong production of extracellular functional amyloid (FuBA) and manganese oxidation. This is the first completed genome sequence of the family Geodermatophilaceae. The 5,322,497 bp long genome with its 5,161 protein-coding and 58 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Similar content being viewed by others
Introduction
Strain G-20T (= DSM 43160 = ATCC 25078 = JCM 3152) is the type strain of the species Geodermatophilus obscurus, which is the type genus in the family Geodermatophilaceae [1,2]. The species name derives from the Latin word ‘obscurus’ meaning dark, obscure, indistinct, unintelligible [1]. The genus Geodermatophilus and family Geodermatophilaceae were originally proposed in 1968 by Luedemann [1]. The genus Geodermatophilus was first described as a genus closely related to genus Dermatophilus, but being isolated from soil, as indicated by the prefix ‘geo’, which derives from Greek ‘Gea’ meaning Earth [1]. In contrast, members of the genus Dermatophilus originated from skin lesions of cattle, sheep, horses, deer, and man [3], as the meaning of the genus name is ‘skin-loving’. Yet, on the basis of 16S rRNA gene sequences, Geodermatophilus proved to be only distantly related to Dermatophilus [4] and was thus included in 1989 in the family Frankiaceae [5], together with the genera Blastococcus and Frankia. In 1996, the genera Dermatophilus and Blastococcus were excluded again from the family Frankiaceae [6] and finally formally combined with the genus Modestobacter in the family Geodermatophilaceae again [2]. G. obscurus is the only validly described species in the genus Geodermatophilus [7], and consists of four subspecies [1] which have never been validly published [8].
The type strain G-20T, together with other strains, has been isolated from soil in the Amargosa Desert of Nevada, USA [3]. Further Geodermatophilus strains were isolated from limestone [8,9] and rock varnish [10] in the Negev Desert, Israel, from marble in Delos, Greece [8,9], from chestnut soil in Gardabani, Central Georgia [11], from rock varnish in the Whipple Mountains, California, USA [12], from orange patina of calcarenite in Noto, Italy [13], from gray to black patinas on marble in Ephesus, Turkey [13], and from high altitude Mount Everest soils [14,15]. Here we present a summary classification and a set of features for G. obscurus G-20T, together with the description of the complete genomic sequencing and annotation.
Classification and features
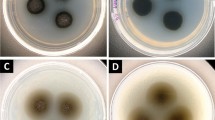
Cells of Geodermatophilus produce densely packed cell aggregates [8], which are described as a muriform, tuber-shaped, noncapsulated, holocarpic thallus consisting of masses of cuboid cells averaging 0.5 to 2.0 µm in diameter (Table 1 and Figure 1) [1]. The thallus breaks up, liberating cuboid or coccoid nonmotile cells and elliptical to lanceolate zoospores [1]. The single cell can differentiate further into polar flagellated motile zoospores [15]. Thus, cells of Dermatophilus may express a morphogenetic growth cycle in which it switches between a thalloid C-form and a motile zoosporic R-form [15]. It has been supposed that tryptose (Difco) contains an unidentified factor, M, which controls morphogenesis in Geodermatophilus [15], though others could not observe the motile, budding zoospores of the R-form [8]. As colonies, strains of Geodermatophilus strains exhibit usually a dark brownish, greenish, or black pigmentation with a smooth to rough surface and in most cases a solid consistency, including minor variations in colony shape [8]. Young colonies are almost colorless, having smooth edges which become distorted and lobed in older colonies, where the colony consistency becomes somewhat crumby [8]. The colonies become darkly pigmented immediately when they started to protrude upwards in the space above the agar [8]. Geodermatophilus does not produce hyphae, vesicles, outer membranous spore layers or capsules [5].
Strain G-20T utilizes l-arabinose, d-galactose, d-glucose, glycerol, inositol, d-levulose, d-mannitol, sucrose, and d-xylose as single carbon sources for growth, but not d-arabinose, dulcitol, β-lactose, melezitose, α-melibiose, raffinose, D-ribose, and ethanol [1,23]. Growth with l-rhamnose is only poor [1]. Strain G-20T is negative for β-hemolysis of blood agar (10% human blood) [1]. Also, nitrate reduction occurs only sporadically with both inorganic or organic nitrate broth [1]. Strain G-20T hydrolyses starch, is weakly positive for gelatin liquefaction and negative for casein utilization [23].
Strain G-20T showed a remarkable production of extracellular functional bacterial amyloid (FuBA), which is accessible to WO2 antibodies without saponification [24]. The WO2 antibody has been shown to bind only to amyloid and not to other kinds of protein aggregates [20,24]. One strain of G. obscurus was described as having a lytic activity on yeast cell walls [12]. Another strain from rock varnish was shown to exhibit very strong resistance to UV-C light (220 J×m-2) [12]. Two strains from rock varnish in the Negev Desert were able to oxidize manganese [10].
Only three G. obscurus isolates have 16S rRNA gene sequences with >98% sequence similarity to strain G-20T: isolate G18 from Namibia, 99.1% [2], isolate 06102S3-1 from deep-sea sediments of the East Pacific and Indian Ocean (EU603760) 98.5%, and G. obscurus subspecies utahensis DSM 43162, 98.03% [8]. The highest degree of sequence similarity in environmental metagenomic surveys, 93.3% was reported from a marine metagenome (AACY020064011) from the Sargasso Sea [25]. (January 2010).
Figure 2 shows the phylogenetic neighborhood of for G. obscurus G-20T in a 16S rRNA based tree. The sequences of the three 16S rRNA gene copies in the genome of G. obscurus G-20T do not differ from each other, but differ by 24 nucleotides from the previously published 16S rRNA sequence obtained from DSM 43160 (X92356). These considerable discrepancies are most likely due to sequencing errors in the latter sequence. Genbank accession L40620, which was obtained from ATCC 25078, differs by only one single nucleotide from the 16S rRNA gene copies in the genome obtained from DSM 43160.
Phylogenetic tree highlighting the position of G. obscurus G-20T relative to the other type strains within the suborder Frankineae. The tree was inferred from 1,364 aligned characters [26,27] of the 16S rRNA gene sequence under the maximum likelihood criterion [28] and rooted with the type strain of the order Actinomycetales. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 350 bootstrap replicates [29] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [30], such as the GEBA organism Nakamurella multipartita [31] are shown in blue. Important non-type strains are shown in green [32], and published genomes in bold.
Chemotaxonomy
The major fatty acids of strain G-20T are iso-C15:0 (19.0%), iso-C16:0 (16.2%), C16:1 cis9 (13:0%), C17:1 (10.4%), C18:1 cis9 (6.6%), and anteiso-C15:0 (5.7%). All other fatty acids (iso-C14:0, C15:0, C15:1, C16:0, C17:0, iso-C17:0, and anteiso-C17:0) were each below 4% [33]. Qualitatively, these values are largely congruent with other sources [4,34]. Strain G-20T contains tetrahydro-menaquinones with nine isoprene units [MK-9(H4)] as sole component [4]. No whole cell wall sugar was found in strain G-20T, which contains only small amounts of galactose, glucose, and ribose [4,35]. The cell wall type is IIIC, and contains meso-2,6-diaminopimelic acid [35].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genome OnLine Database [30] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Growth conditions and DNA isolation
G. obscurus G-20T, DSM 43160, was grown in DSMZ medium 65 [36] at 28°C. DNA was isolated from 0.5–1 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) with a modified protocol for cell lysis, (procedure st/L), and one hour incubation at 37°C, according to Wu et al. [37].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger and 454 sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at the JGI website (http://www.jgi.doe.gov/). 454 Pyrosequencing reads were assembled using the Newbler assembler version 1.1.02.15 (Roche). Large Newbler contigs were broken into 5,725 overlapping fragments of 1,000 bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the parallel phrap assembler (High Performance Software, LLC). Possible misassemblies were corrected with Dupfinisher or transposon bombing of bridging clones [38]. A total of 1,530 Sanger finishing reads were produced to close gaps, to resolve repetitive regions, and to raise the quality of the finished sequence. Illumina reads were used to improve the final consensus quality using an in-house developed tool (the Polisher). The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Sanger and 454 sequencing platforms provided 29.8× coverage of the genome. The final assembly contains 48,209 Sanger reads and 353,553 pyrosequencing reads.
Genome annotation
Genes were identified using Prodigal [39] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [40]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [41].
Genome properties
The genome is 5,322,497 bp long and comprises one main chromosome with a 74.0% GC content (Table 3 and Figure 3). Of the 5,219 genes predicted 5,161 were protein coding genes, and 58 RNAs. In addition, 350 pseudogenes were also identified. The majority of the protein-coding genes (69.8%) were assigned with a putative function while those remaining were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Comparison with closest related genomes
Table 5 provides an overall comparison of the genomes of G. obscurus strain G-20T with the closest available genomes, that is, Acidothermus cellulolyticus 11BT, Frankia alni ACN14A and N. multipartita Y-104T. The total length of (non-overlapping) high-scoring segment pairs (HSPs) and the number of identical base pairs within these HSPs were determined using the GGDC web server [42] by directly applying NCBI Blastn to the genomes represented as nucleotide sequences [43].
Number and proportion of shared homologs were determined using the ‘Phylogenetic Profiler’ function of the IMG system [41] using default values. While the relative order of 16S rRNA difference does not correspond to the genomic similarities, the four genome-based measures uniformly indicate that N. multipartita Y-104T possesses the genome most similar to the one of G. obscurus G-20T, followed by F. alni ACN14A and A. cellulolyticus 11BT.
References
Luedemann GM. Geodermatophilus, a new genus of the Dermatophilaceae (Actinomycetales). J Bacteriol 1968; 96:1848–1858. P
Normand P. Geodermatophilaceae fam. nov., a formal description. Int J Syst Evol Microbiol 2006; 56:2277–2278. PubMed doi:10.1099/ijs.0.64298-0
Gordon MA. The genus Dermatophilus. J Bacteriol 1964; 88:509–522. PubMed
Stackebrandt E, Kroppenstedt RM, Fowler VJ. A phylogenetic analysis of the family Dermatophilaceae. J Gen Microbiol 1983; 129:1831–1838. PubMed
Hahn D, Lechevalier MP, Fischer A, Stackebrandt E. Evidence for a close phylogenetic relationship between members of the genera Frankia, Geodermatophilus and Blastococcus and emendation of the family Frankiaceae. Syst Appl Microbiol 1989; 11:236–242.
Normand P, Orso S, Cournoyer B, Jeannin P. Chapelon, Dawson J, Evtushenko L, Misra AK. Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol 1996; 46:1–9. PubMed doi:10.1099/00207713-46-1-1
Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzéby J, Tindall BJ. Taxonomic outline of the Bacteria and Archaea, Release 7.7 March 6, 2007. Part 10 — The Bacteria: Phylum Actinobacteria: Class “Actinobacteria”. http://www.taxonomicoutline.org
Eppard M, Krumbein WE, Koch C, Rhiel E, Staley JT, Stackebrandt E. Morphological, physiological, and molecular characterization of actinomycetes isolated from dry soil, rocks, and monument surfaces. Arch Microbiol 1996; 166:12–22. PubMed doi:10.1007/s002030050350
Salazar O, Valverde A, Genilloud O. Real-Time PCR for the detection and quantification of Geodermatophilaceae from stone samples and identification of new members of the genus Blastococcus. Appl Environ Microbiol 2006; 72:346–352. PubMed doi:10.1128/AEM.72.1.346-352.2006
Hungate B, Danin A, Pellerin NB, Stemmler J, Kjellander P, Adams JB, Staley JT. Characterization of manganese-oxidizing (MnII->MnIV) bacteria from Negev Desert rock varnish: implications in desert varnish formation. Can J Microbiol 1987; 33:939–943.
Kudukhashvili PG, Gurielidze MA, Pataraya DT. Study of the lytic activities of actinomycetes isolated from different soils in Georgia. Appl Biochem Microbiol 2001; 37:251–252. doi:10.1023/A:1010272900216
Kuhlman KR, Allenbach LB, Ball CL, Fusco WG, La Duc MT, Kuhlman GM, Anderson RC, Stuecker T, Erickson IK, Benardini J, et al. Enumeration, isolation, and characterization of ultraviolet (UVC) resistant bacteria from rock varnish in the Whipple Mountains, California. Icarus 2005; 174:585–595. doi:10.1016/j.icarus.2004.11.022
Urzì C, Brusetti L, Salamone P, Sorlini C, Stackebrandt E, Daffonchio D. Biodiversity of Geodermatophilaceae isolated from altered stones and monuments in the Mediterranean basin. Environ Microbiol 2001; 3:471–479. PubMed doi:10.1046/j.1462-2920.2001.00217.x
Ishiguro EE, Fletcher DW. Characterization of Geodermatophilus strains isolated from high altitude Mount Everest soils. Mikrobiologika 1975; 12:99–108.
Ishiguro EE, Wolfe RS. Control of morphogenesis in Geodermatophilus: ultrastructural studies. J Bacteriol 1970; 104:566–580. PubMed
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed doi:10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed doi:10.1073/pnas.87.12.4576
Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Springer, New York, 2001, p. 119–169.
Stackebrandt E. Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479–491. doi:10.1099/00207713-47-2-479
O’Nuallain B, Wetzel R. Conformational Abs recognizing a generic amyloid fibril epitope. Proc Natl Acad Sci USA 2002; 99:1485–1490. PubMed doi:10.1073/pnas.022662599
Classification of Bacteria and Archaea in risk groups www.baua.de TRBA_466.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25–29. PubMed doi:10.1038/75556
Mevs U, Stackebrandt E, Schumann P, Gallikowski C, Hirsch P. Modestobacter multiseptatus gen. nov., sp. nov., a budding actinomycete from soils of the Asgard Range (Transantarctic Mountains). Int J Syst Evol Microbiol 2000; 50:337–346. PubMed
Jordal PB, Dueholm MS, Larsen P, Petersen SV, Enghild JJ, Christiansen G, Hojrup P, Nielsen PH, Otzen DE. Widespread abundance of functional bacterial amyloid in mycolata and other grampositive bacteria. Appl Environ Microbiol 2009; 75:4101–4110. PubMed doi:10.1128/AEM.02107-08
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004; 304:66–74. PubMed doi:10.1126/science.1093857
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540–552. PubMed
Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452–464. PubMed doi:10.1093/bioinformatics/18.3.452
Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 2008; 57:758–771. PubMed doi:10.1080/10635150802429642
Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How Many Bootstrap Replicates Are Necessary? Lect Notes Comput Sci 2009; 5541:184–200. doi:10.1007/978-3-642-02008-7_13
Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346–D354. PubMed doi:10.1093/nar/gkp848
Tice H, Mayilraj S, Sims D, Lapidus A, Nolan M, Lucas S, Glavina Del Rio T, Copeland A, Cheng JF, Meincke L, et al. Complete genome sequence of Nakamurella multipartita type strain (Y-104T). Stand Genomic Sci 2010; 2:168–175.
Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N, et al. Cenome characteristics of facultatively symbiontic Frankia sp. strains reflect host range and host plant biogeography. Genome Res 2007; 17:7–15. PubMed doi:10.1101/gr.5798407
Mirza MS, Janse JD, Hahn D, Akkermans ADL. Identification of atypical Frankia strains by fatty acid analysis. FEMS Microbiol Lett 1991; 83:91–98. doi:10.1111/j.1574-6968.1991.tb04395.x
Wink JM. Compendium of Actinobacteria.
Lechevalier MP, Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 1970; 20:435–443. doi:10.1099/00207713-20-4-435
List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/media_list.php
Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova N, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462:1056–1060. PubMed doi:10.1038/nature08656
Sims D, Brettin T, Detter J, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12–20. doi:10.4056/sigs.761
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. BMC Bioinformatics 2010; 11:119. PubMed doi:10.1186/1471-2105-11-119
Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. Nat Methods (In press).
Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271–2278. PubMed doi:10.1093/bioinformatics/btp393
Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring sequence pairs. Stand Genomic Sci 2010; 2:142–148. doi:10.4056/sigs.541628
Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2010; 2:117–134. doi:10.4056/sigs.531120
Acknowledgements
We would like to gratefully acknowledge the help of Susanne Schneider (DSMZ) for DNA extraction and quality analysis. This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-1 and SI 1352/1-2.
Author information
Authors and Affiliations
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Ivanova, N., Sikorski, J., Jando, M. et al. Complete genome sequence of Geodermatophilus obscurus type strain (G-20T). Stand in Genomic Sci 2, 158–167 (2010). https://doi.org/10.4056/sigs.711311
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.711311