Abstract
Ensifer meliloti strain RRI128 is an aerobic, motile, Gram-negative, non-spore-forming rod. RRI128 was isolated from a nodule recovered from the roots of barrel medic (Medicago truncatula) grown in the greenhouse and inoculated with soil collected from Victoria, Australia. The strain is used in commercial inoculants in Australia. RRI128 nodulates and forms an effective symbiosis with a diverse range of lucerne cultivars (Medicago sativa) and several species of annual medic (M. truncatula, Medicago littoralis and Medicago tornata), but forms an ineffective symbiosis with Medicago polymorpha. Here we describe the features of E. meliloti strain RRI128, together with genome sequence information and annotation. The 6,900,273 bp draft genome is arranged into 156 scaffolds of 157 contigs, contains 6,683 protein-coding genes and 87 RNA-only encoding genes, and is one of 100 rhizobial genomes sequenced as part of the DOE Joint Genome Institute 2010 Genomic Encyclopedia for Bacteria and Archaea-Root Nodule Bacteria (GEBA-RNB) project.
Similar content being viewed by others
Introduction
Ensifer meliloti strain RRI128 is used in Australia to produce commercial peat cultures (referred to as Group AL inoculants) mainly for the inoculation of lucerne (Medicago sativa L.). Lucerne is sown on about 600, 000 ha annually (A. Humphries pers. com.) and is nearly always inoculated prior to sowing. RRI128 is also used for the inoculation of strand medic (Medicago littoralis Loisel) and disc medic (Medicago tornata (L.) Miller), a hybrid of the two former species, and bokhara clover (Melilotus albus Medik). RRI128 has been used commercially since 2000 when it replaced strain WSM826 [1]. Strain RRI128 was isolated from a nodule from the roots of barrel medic (Medicago truncatula Gaertn) growing in the greenhouse and inoculated with an alkaline sandy soil (pHCaCl2 7.6) collected by J. Slattery, near Tempy, Victoria.
The strain was selected for use in commercial inoculants following assessment of its nitrogen fixation capacity (effectiveness), growth on acidified agar and saprophytic competence in an in-situ soil study [2], with supporting data of satisfactory performance at ten field sites. Additional testing has shown RRI128 to be effective on 28 cultivars of lucerne (Ballard unpub. data). It also forms effective symbiosis with a range of strand and disc medics [2] which show symbiotic affinity with lucerne [3,4].
Soil acidity has long been recognized as a constraint to lucerne nodulation [5] with some evidence that strains of E. meliloti have less acidity tolerance than Ensifer medicae, possibly due to their association with Medicago species that favor neutral to alkaline soils [6]. With RRI128, constraints to lucerne nodulation are observed around pH 5. Nodulation of lucerne seedlings inoculated with RRI128 was 42% at pH 5.0 in solution culture experiments [7] and observed to decline rapidly at field sites where pHCaCl2 was below 4.7 (Ballard, unpub. data). Other strains (e.g. SRDI672) have increased lucerne nodulation in solution culture at pH 4.8 (61% cf. 12% of lucerne seedlings with nodules) but are probably approaching the limit of acidity tolerance for E. meliloti [8].
Stable colony morphology and cell survival on seed make RRI128 amenable to commercial use. RRI128 produces colonies of consistent appearance and with moderate polysaccharide when grown on yeast mannitol agar, enabling easy visual assessment of culture purity. It differs in this regard from the strain it replaced (WSM826) which produced ‘dry’ and ‘mucoid’ colony variants, in common with many of the strains that nodulate lucerne and medic [9]. When applied correctly RRI128 has been shown to survive at more than 10,000 cells per lucerne seed at six weeks after inoculation [10]. Good survival may well be characteristic of E. meliloti, since former inoculant strain WSM826 is equally competent in this regard [11,12].
Here we present a preliminary description of the general features of E. meliloti strain RRI128 together with its genome sequence and annotation.
Classification and general features
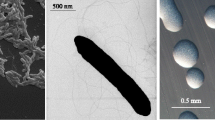
Ensifer meliloti strain RRI128 is a motile, non-sporulating, non-encapsulated, Gram-negative rod in the order Rhizobiales of the class Alphaproteobacteria. The rod-shaped form varies in size with dimensions of approximately 0.5 µm in width and 1.0–2.0 µm in length (Figure 1A). It is fast growing, forming colonies within 3–4 days when grown on TY [13] or half strength Lupin Agar (½LA) [14] at 28°C. Colonies on ½LA are opaque, slightly domed and moderately mucoid with smooth margins (Figure 1B).
Minimum Information about the Genome Sequence (MIGS) is provided in Table 1. Figure 2 shows the phylogenetic neighborhood of Ensifer meliloti strain RRI128 in a 16S rRNA gene sequence based tree. This strain has 100% sequence identity (1366/1366 bp) at the 16S rRNA sequence level to the fully sequenced E. meliloti Sm1021 [30] and 99% 16S rRNA sequence (1362/1366 bp) identity to the fully sequenced E. medicae strain WSM419 [31].
Phylogenetic tree showing the relationship of Ensifer meliloti strain RRI128 (shown in bold) with some of the root nodule bacteria in the order Rhizobiales based on aligned sequences of the 16S rRNA gene (1,307 bp internal region). All sites were informative and there were no gap-containing sites. Phylogenetic analyses were performed using MEGA [27], version 5.05. The tree was built using the maximum likelihood method with the General Time Reversible model. Bootstrap analysis [28] with 500 replicates was performed to assess the support of the clusters. Type strains are indicated with a superscript T. Brackets after the strain name contain a DNA database accession number and/or a GOLD ID (beginning with the prefix G) for a sequencing project registered in GOLD [29]. Published genomes are indicated with an asterisk.
Symbiotaxonomy
Ensifer meliloti strain RRI128 forms nodules on (Nod+) and fixes N2 (Fix+) with Medicago sativa, Melillotus albus and Trigonella balansae (Boiss. and Reuter). It also forms effective symbiosis with several species of annual medic (M. truncatula, M. littoralis and M. tornata) that happen to be closely related to each other based on their ability to be hybridized [5] and morphological and nucleotide sequence analyses of their relatedness [32]. RRI128 forms ineffective (white) nodules with Medicago polymorpha, a species that is generally recognized to have a more specific rhizobial requirement for effective symbiosis than Medicago sativa and Medicago littoralis [4,33] (Table 2).
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its environmental and agricultural relevance to issues in global carbon cycling, alternative energy production, and biogeochemical importance, and is part of the Community Sequencing Program at the U.S. Department of Energy, Joint Genome Institute (JGI) for projects of relevance to agency missions. The genome project is deposited in the Genomes OnLine Database [29] and an improved-high-quality-draft genome sequence in IMG/GEBA. Sequencing, finishing and annotation were performed by the JGI. A summary of the project information is shown in Table 3.
Growth conditions and DNA isolation
Ensifer meliloti strain RRI128 was cultured to mid logarithmic phase in 60 ml of TY rich medium on a gyratory shaker at 28°C [35]. DNA was isolated from the cells using a CTAB (Cetyl trimethyl ammonium bromide) bacterial genomic DNA isolation method [36].
Genome sequencing and assembly
The genome of Ensifer meliloti strain RRI128 was sequenced at the Joint Genome Institute (JGI) using Illumina [37] technology. An Illumina standard shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform, which generated 13,085,546 reads totaling 1,962 Mb of Illumina data.
All general aspects of library construction and sequencing performed at the JGI can be found at the JGI user home [36]. All raw Illumina sequence data was passed through DUK, a filtering program developed at JGI, which removes known Illumina sequencing and library preparation artifacts (Mingkun, L., Copeland, A. and Han, J., unpublished). The following steps were then performed for assembly: (1) filtered Illumina reads were assembled using Velvet [38], version 1.1.04, (2) 1–3 Kb simulated paired end reads were created from Velvet contigs using wgsim [39], (3) Illumina reads were assembled with simulated read pairs using Allpaths-LG [40] (version r39750).
Parameters for assembly steps were:
-
(1)
Velvet (Velvet optimizer params: —v —s 51 —e 71 —i 2 —t 1 —f “-shortPaired -fastq $FASTQ” —o “-ins_length 250 -min_contig_lgth 500”)
-
(2)
wgsim (-e 0 -1 76 -2 76 -r 0 -R 0 -X 0,) (3) Allpaths-LG (PrepareAllpathsInputs:PHRED64=1 PLOIDY=1 FRAGCOVERAGE=125 JUMPCOVERAGE=25 LONGJUMPCOV=50, RunAllpath-sLG: THREADS=8 RUN=stdshredpairs TARGETS=standard VAPIWARNONLY=True OVERWRITE=True).
The final draft assembly contained 157 contigs in 156 scaffolds. The total size of the genome is 6.9 Mb and the final assembly is based on 1,962 Mb of Illumina data, which provides an average 285× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [41] as part of the Oak Ridge National Laboratory genome annotation pipeline. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. These data sources were combined to assert a product description for each predicted protein. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [42] RNAMMer [43], Rfam [44], TMHMM [45], and SignalP [46]. Additional gene prediction analyses and functional annotation were performed within the Integrated Microbial Genomes (IMG-ER) platform [47].
Genome properties
The genome is 6,900,273 nucleotides with 61.98% GC content (Table 4) and comprised of 156 scaffolds (Figures 3a,3b,3c,3d,3e). From a total of 6,770 genes, 6,683 were protein encoding and 87 RNA only encoding genes. The majority of genes (78.79%) were assigned a putative function whilst the remaining genes were annotated as hypothetical. The distribution of genes into COGs functional categories is presented in Table 5.
Graphical map of YU7DRAFT_scaffold_0.1 of the genome of Ensifer meliloti strain RRI128. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical map of YU7DRAFT_scaffold_1.2 of the genome of Ensifer meliloti strain RRI128. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical map of YU7DRAFT_scaffold_2.3 of the genome of Ensifer meliloti strain RRI128. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical map of YU7DRAFT_scaffold_3.4 of the genome of Ensifer meliloti strain RRI128. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical map of YU7DRAFT_scaffold_4.5 of the genome of Ensifer meliloti strain RRI128. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
References
Bullard GK, Roughley RJ, Pulsford DJ. The legume inoculant industry and inoculant quality control in Australia: 1953–2003. Aust J Exp Agric 2005; 45:127–140. http://dx.doi.org/10.1071/EA03159
Ballard RA, Slattery JF, Charman N. Host range and saprophytic competence of Sinorhizobium meliloti — a comparison of strains for the inoculation of lucerne, strand and disc medics. Aust J Exp Agric 2005; 45:209–216. http://dx.doi.org/10.1071/EA03126
Howieson JG, Nutt B, Evans P. Estimation of host-strain compatibility for symbiotic N-fixation between Rhizobium meliloti, several annual species of Medicago and Medicago sativa. Plant Soil 2000; 219:49–55. http://dx.doi.org/10.1023/A:1004795617375
Brockwell J, Hely FW. Symbiotic characteristics of Rhizobium meliloti: an appraisal of the systematic treatment of nodulation and nitrogen fixation interactions between hosts and rhizobia of diverse origins. Australian Journal of Agricultural Economics 1966; 17:885–889.
Munns DN. Nodulation of Medicago sativa in solution culture. 1. Acid-sensitive steps. Plant Soil 1968; 28:129–146. http://dx.doi.org/10.1007/BF01349181
Garau G, Reeve WG, Brau L, Yates RJ, James D, Tiwari R, O’Hara GW, Howieson JG. The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil 2005; 276:263–277. http://dx.doi.org/10.1007/s11104-005-0374-0
Charman N, Ballard RA, Humphries AW, Auricht GC. Improving lucerne nodulation at low pH: contribution of rhizobial and plant genotype to the nodulation of lucerne seedlings growing in solution culture at pH 5. Aust J Exp Agric 2008; 48:512–517. http://dx.doi.org/10.1071/EA07138
Charman N, Humphries A, Smith D, Drew E, Marshall E, Denton M, Hayes R, Venkatanagappa S, Ballard R. Selection of lucerne and rhizobia for improved nodulation at low pH. 15th Australian Nitrogen Fixation Conference Margaret River, Western Australia 2009. p 49.
McInnes A, Holford P, Thies JE. Characterisation of dry and mucoid colonies isolated from Australian rhizobial inoculant strains for Medicago species. Anim Prod Sci 2005; 45:151–159. http://dx.doi.org/10.1071/EA03125
Garau G, Yates RJ, Deiana P, Howieson JG. Novel strains of nodulating Burkholderia have a role in nitrogen fixation with Papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem 2009; 41:125–134. http://dx.doi.org/10.1016/j.soilbio.2008.10.011
Gemell LG, Hartley EJ, Herridge DF. Point-of-sale evaluation of preinoculated and custom-inoculated pasture legume seed. Aust J Exp Agric 2005; 45:161–169. http://dx.doi.org/10.1071/EA03151
Hartley E, Gemell G, Deaker R. Some factors that contribute to poor survival of rhizobia on preinoculated legume seed. Crop Pasture Sci 2012; 63:858–865. http://dx.doi.org/10.1071/CP12132
Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 1974; 84:188–198. PubMed http://dx.doi.org/10.1099/00221287-84-1-188
Howieson JG, Ewing MA, D’antuono MF. Selection for acid tolerance in Rhizobium meliloti. Plant Soil 1988; 105:179–188. http://dx.doi.org/10.1007/BF02376781
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen M, Angiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed http://dx.doi.org/10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed http://dx.doi.org/10.1073/pnas.87.12.4576
Chen WX, Wang ET, Kuykendall LD. The Proteobacteria. New York: Springer-Verlag; 2005.
Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. In: Garrity GM, Brenner DJ, Kreig NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Second ed: New York: Springer-Verlag; 2005. p 1.
Kuykendall LD. Order VI. Rhizobiales ord. nov. In: Garrity GM, Brenner DJ, Kreig NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Second ed: New York: Springer-Verlag; 2005. p 324.
Kuykendall LD. Family I. Rhizobiaceae In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. New York: Springer-Verlag; 2005. p 324.
Young JM. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination “Sinorhizobium adhaerens” (Casida 1982) Willems et al. 2003 legitimate? Request for an Opinion. Int J Syst Evol Microbiol 2003; 53:2107–2110. PubMed http://dx.doi.org/10.1099/ijs.0.02665-0
Judicial Commission of the International Committee on Systematics of P. The genus name Sinorhizobium Chen et al. 1988 is a later synonym of Ensifer Casida 1982 and is not conserved over the latter genus name, and the species name ‘Sinorhizobium adhaerens’ is not validly published. Opinion 84. International Journal of Systematic and Evolutionary Microbiology 2008;58(Pt 8):1973.
Rome S, Brunel B, Normand P, Fernandez M, Cleyet-Marel JC. Evidence that two genomic species of Rhizobium are associated with Medicago truncatula. Arch Microbiol 1996; 165. PubMed http://dx.doi.org/10.1007/s002030050328
Rome S, Cleyet-Marel JC, Materon LA, Normand P, Brunel B. Rapid identification of Medicago nodulating strains by using two oligonucleotide probes complementary to 16S rDNA sequences. Can J Microbiol 1997; 43:854–861. PubMed http://dx.doi.org/10.1139/m97-124
Biological Agents. Technical rules for biological agents. TRBA (http://www.baua.de):466.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. PubMed http://dx.doi.org/10.1038/75556
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, evolutionary distance, and Maximum Parismony methods. Mol Biol Evol 2011; 28:2731–2739. PubMed http://dx.doi.org/10.1093/molbev/msr121
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39:783–791. http://dx.doi.org/10.2307/2408678
Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475–D479. PubMed http://dx.doi.org/10.1093/nar/gkm884
Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 2001; 293:668–672. PubMed http://dx.doi.org/10.1126/science.1060966
Reeve W, Chain P, O’Hara G, Ardley J, Nandesena K, Brau L, Tiwari R, Malfatti S, Kiss H, Lapidus A, et al. Complete genome sequence of the Medicago microsymbiont Ensifer (Sinorhizobium) medicae strain WSM419. Stand Genomic Sci 2010; 2:77–86. PubMed http://dx.doi.org/10.4056/sigs.43526
Small E. Alfalfa and Relatives: Evolution and Classification of Medicago. Ottawa, Canada: NRC Reserach Press; 2011.
Ballard RA, Charman N. Nodulation and growth of pasture legumes with naturalised soil rhizobia. 1. Annual Medicago spp. Aust J Exp Agric 2000; 40:939–948. http://dx.doi.org/10.1071/EA99112
Howieson JH, Ballard RA, deKoning C, Sandral G, Charman N. Trigonella balansae - a new pasture legume for the alkaline soils of southern Australia. 10th Australian Agronomy Conference. Hobart, Australia.: Australian Society of Agronomy; 2001.
Reeve WG, Tiwari RP, Worsley PS, Dilworth MJ, Glenn AR, Howieson JG. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology 1999; 145:1307–1316. PubMed http://dx.doi.org/10.1099/13500872-145-6-1307
JGI user home. http://my.jgi.doe.gov/general/index.html
Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433–438. PubMed http://dx.doi.org/10.1517/14622416.5.4.433
Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Current Protocols in Bioinformatics 2010;Chapter 11:Unit 11 5.
wgsim. https://github.com/lh3/wgsim
Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA 2011; 108:1513–1518. PubMed http://dx.doi.org/10.1073/pnas.1017351108
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119. PubMed http://dx.doi.org/10.1186/1471-2105-11-119
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955–964. PubMed
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100–3108. PubMed http://dx.doi.org/10.1093/nar/gkm160
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res 2003; 31:439–441. PubMed http://dx.doi.org/10.1093/nar/gkg006
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567–580. PubMed http://dx.doi.org/10.1006/jmbi.2000.4315
Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783–795. PubMed http://dx.doi.org/10.1016/j.jmb.2004.05.028
Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271–2278. PubMed http://dx.doi.org/10.1093/bioinformatics/btp393
Acknowledgements
This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. We gratefully acknowledge the funding received from the Murdoch University Strategic Research Fund through the Crop and Plant Research Institute (CaPRI) and the Centre for Rhizobium Studies (CRS) at Murdoch University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Reeve, W., Ballard, R., Drew, E. et al. Genome sequence of the Medicago-nodulating Ensifer meliloti commercial inoculant strain RRI128. Stand in Genomic Sci 9, 602–613 (2014). https://doi.org/10.4056/sigs.4929626
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.4929626