Abstract
Granulicella tundricola strain MP5ACTX9T is a novel species of the genus Granulicella in subdivision 1 Acidobacteria. G. tundricola is a predominant member of soil bacterial communities, active at low temperatures and nutrient limiting conditions in Arctic alpine tundra. The organism is a cold-adapted acidophile and a versatile heterotroph that hydrolyzes a suite of sugars and complex polysaccharides. Genome analysis revealed metabolic versatility with genes involved in metabolism and transport of carbohydrates, including gene modules encoding for the carbohydrate-active enzyme (CAZy) families for the breakdown, utilization and biosynthesis of diverse structural and storage polysaccharides such as plant based carbon polymers. The genome of G. tundricola strain MP5ACTX9T consists of 4,309,151 bp of a circular chromosome and five mega plasmids with a total genome content of 5,503,984 bp. The genome comprises 4,705 protein-coding genes and 52 RNA genes.
Similar content being viewed by others
Introduction
The strain MP5ACTX9T (=ATCC BAA-1859T =DSM 23138T) is the type strain of Granulicella tundricola [tun.dri.co’la. N.L. n. tundra, tundra, a cold treeless region; L. masc. suffix-cola (from L. n. incola) dweller; N.L. n. tundricola tundra dweller] that was isolated from soil at the Malla Nature Reserve, Kilpisjärvi, Finland; 69°01′N, 20°50′E) and described along with other species of the genus Granulicella isolated from tundra soil [1].
Acidobacteria is a phylogenetically and physiologically diverse phylum [2,3], the members of which are ubiquitously found in diverse habitats and are abundant in most soil environments [4,5] including Arctic tundra soils [6,7]. Acidobacteria are relatively difficult to cultivate, as they have slow growth rates. To date only subdivisions 1, 3, 4, 8, 10 and 23 Acidobacteria are defined by taxonomically characterized representatives [8–23] as well as three ‘Candidatus’ taxa [24,25]. The phylogenetic diversity, ubiquity and abundance of this group suggest that they play important ecological roles in soils. The abundance of Acidobacteria correlates with soil pH [26,27] and carbon [28,29], with subdivision 1 Acidobacteria being most abundant in slightly acidic soils. Acidobacteria, including members of the genera Granulicella and Terriglobus, dominate the acidic tundra heaths of northern Finland [26,30–32]. Using selective isolation techniques we have been able to isolate several slow growing and fastidious strains of Acidobacteria [1,11]. On the basis of phylogenetic, phenotypic and chemotaxonomic data, including 16S rRNA, rpoB gene sequence similarity and DNA-DNA hybridization, strain MP5ACTX9T was classified as a novel species of the genus Granulicella [1]. Here, we summarize the physiological features together with the complete genome sequence, annotation and data analysis of Granulicella tundricola strain MP5ACTX9T.
Classification and features
Within the genus Granulicella, eight species are described with validly published names: G. mallensis MP5ACTX8T, G. tundricola MP5ACTX9T, G. arctica MP5ACTX2T,G. sapmiensis S6CTX5AT isolated from Arctic tundra soil [1] and G. paludicola OB1010T, G. paludicola LCBR1, G. pectinivorans TPB6011T,G. rosea TPO1014T,G. aggregans TPB6028T isolated from sphagnum peat bogs [2].
Strain MP5ACTX9T shares 95.5–97.2% 16S rRNA gene identity with tundra soil strains G. mallensis MP5ACTX8T (95.5%), G. arctica MP5ACTX2T (96.9%), G. sapmiensis S6CTX5AT (97.2%) and 95.2 – 97.7% identity with the sphagnum bog strains, G. pectinivorans TPB6011T (97.7%), G. rosea TPO1014T (97.2%), %), G. aggregans TPB6028T (96.8%), G. paludicola LCBR1 (95.9%), and G. paludicola strain OB1010T (95.3%), which were isolated from sphagnum peat. Phylogenetic analysis based on the 16S rRNA gene of taxonomically classified strains of family Acidobacteriaceae placed G. rosea type strain T4T (AM887759) as the closest taxonomically classified relative of G. tundricola strain MP5ACTX9T (Table 1, Figure 1).
Phylogenetic tree highlighting the position of G. tundricola MP5ACTX9T (shown in bold) relative to the other type strains within subdivision1 Acidobacteria. The maximum likelihood tree was inferred from 1,361 aligned positions of the 16S rRNA gene sequences and derived based on the Tamura-Nei model using MEGA 5 [42]. Bootstrap values >50 (expressed as percentages of 1,000 replicates) are shown at branch points. Bar: 0.01 substitutions per nucleotide position. The corresponding GenBank accession numbers are displayed in parentheses. Strains whose genomes have been sequenced, are marked with an asterisk; G. mallensis MP5ACTX8T (CP003130), G. tundricola MP5ACTX9T (CP002480), T. saanensis SP1PR4T (CP002467), T. roseus KBS63T (CP003379), and A. capsulatum ATCC 51196T (CP001472). Bryobacter aggregatus MPL3 (AM162405) in SD3 Acidobacteria was used as an outgroup.
Morphology and physiology
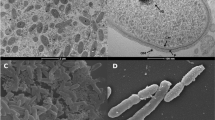
G. tundricola cells are Gram-negative, non-motile, aerobic rods, approximately 0.5 µm wide and 0.5 – 1.8 µm long. Colonies on R2A agar are pink, circular, convex and smooth. Growth occurs at +4 to 28°C and at pH 3.5–6.5 with an optimum at 21–24°C and pH 5 (Fig. 2). Genotypic analyses, including low rpoB gene sequence similarity and phenotypic characteristics clearly distinguished strain MP5ACTX9T from other Granulicella species/strains, leading us to conclude that MP5ACTX9T represents a novel species of the genus Granulicella, for which the name Granulicella tundricola sp. nov. was proposed [1].
Strain MP5ACTX9T hydrolyzed complex to simple carbon substrates [1] which include complex polysaccharides like aesculin, pectin, laminarin, starch and pullulan, but not gelatin, cellulose, lichenan, sodium alginate, xylan, chitosan or chitin. Strain MP5ACTX9T also utilized the following sugars as growth substrates: D-glucose, maltose, cellobiose, D-fructose, D-galactose, lactose, lactulose, D-mannose, sucrose, trehalose, D-xylose, raffinose, N-acetyl-D-glucosamine, glutamate and gluconic acid. Enzyme activities reported for the strain MP5ACTX9T include acid phosphatase, esterase (C4 and C8), leucine arylamidase, valine arylamidase, α-chymotrypsin, trypsin, naphthol-AS-BI-phosphohydrolase, α- and β-galactosidases, α- and β-glucosidases, N-acetyl-β-glucosaminidase, β-glucuronidase, α-fucosidase and α-mannosidase but negative for alkaline phosphatase and lipase (C14). Strain MP5ACTX9T is resistant to ampicillin, erythromycin, chloramphenicol, neomycin, streptomycin, tetracycline, gentamicin, bacitracin, polymyxin B and penicillin, but susceptible to rifampicin, kanamycin, lincomycin and novobiocin.
Chemotaxonomy
The major cellular fatty acids in G. tundricola are iso-C15:0 (46.4%), C16:1ω7c (35.0%) and C16:0 (6.6%). The cellular fatty acid composition of strain MP5ACTX9T was similar to that of other Granulicella strains with fatty acids iso-C15:0 and C16:1ω7c being most abundant in all strains. Strain MP5ACTX9T contains MK-8 as the major quinone and also contains 4% of MK-7.
Genome sequencing and annotation
Genome project history
G. tundricola strain MP5ACTX9T was selected for sequencing in 2009 by the DOE Joint Genome Institute (JGI) community sequencing program. The Quality Draft (QD) assembly and annotation were completed on May 24, 2010. The GenBank Date of Release was February 2, 2011. The genome project is deposited in the Genomes On-Line Database (GOLD) [43] and the complete genome sequence of strain MP5ACTX9T is deposited in GenBank (CP002480.1). Table 2 presents the project information and its association with MIGS version 2.0 [44].
Growth conditions and genomic DNA extraction
G. tundricola MP5ACTX9T was cultivated on R2 medium as previously described [1]. Genomic DNA (gDNA) of high sequencing quality was isolated using a modified CTAB method and evaluated according to the Quality Control (QC) guidelines provided by the DOE Joint Genome Institute [45].
Genome sequencing and assembly
The finished genome of G. tundricola MP5ACTX9T (JGI ID 4088693) was generated at the DOE Joint genome Institute (JGI) using a combination of Illumina [46] and 454 technologies [47]. For this genome we constructed and sequenced an Illumina GAii shotgun library which generated 42,620,699 reads totaling 3239 Mb, a 454 Titanium standard library which generated 146,119 reads and three paired end 454 libraries with an average insert size of 9.3 kb which generated 178,757 reads totaling 154.3 Mb of 454 data. All general aspects of library construction and sequencing performed at the JGI can be found at the JGI website [45]. The 454 Titanium standard data and the 454 paired end data were assembled with Newbler, version 2.3. Illumina sequencing data was assembled with Velvet, version 0.7.63 [48]. The 454 Newbler consensus shreds, the Illumina Velvet consensus shreds and the read pairs in the 454 paired end library were integrated using parallel phrap, version SPS - 4.24 (High Performance Software, LLC) [49]. The software Consed [50] was used in the finishing process. The Phred/Phrap/Consed software package [51] was used for sequence assembly and quality assessment in the subsequent finishing process. Illumina data was used to correct potential base errors and increase consensus quality using the software Polisher developed at JGI (Alla Lapidus, unpublished). Possible misassemblies were corrected using gapResolution (Cliff Han, un-published), Dupfinisher [52] or sequencing cloned bridging PCR fragments with sub-cloning. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR (J-F Cheng, unpublished) primer walks. The final assembly is based on 29.1 Mb of 454 draft data which provides an average 20× coverage of the genome and 975 Mb of Illumina draft data which provides an average 274× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [53] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [54]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, (COGs) [55,56], and InterPro. These data sources were combined to assert a product description for each predicted protein. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [57], RNAMMer [58], Rfam [59], TMHMM [60], and signalP [61]. Additional gene prediction analysis and functional annotation were performed within the Integrated Microbial Genomes Expert Review (IMG-ER) platform [62].
Genome properties
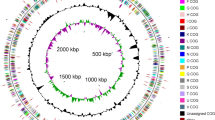
The genome is 5,503,984 bp in size, which includes the 4,309,151 bp chromosome and five plasmids pACIX901 (0.48 Mbp); pACIX902 (0.3 Mbp); pACIX903 (0.19 Mbp), pACIX904 (0.12 Mbp) and pACIX905 (0.12 Mbp), with a GC content of 59.9 mol%. There are 52 RNA genes (Figures 3 and 4, and Table 3). Of the 4,758 predicted genes, 4,706 are protein-coding genes (CDSs) and 163 are pseudogenes. Of the total CDSs, 68.8% represent COG functional categories and 27.5% consist of signal peptides. The distribution of genes into COG functional categories is presented in Figure 3 and Table 4, and Table 5.
Circular representation of the chromosome of G. tundricola MP5ACTX9T displaying relevant genome features. From outside to center; Genes on forward strand (colored by COG categories), genes on reverse strand (colored by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content and GC skew.
Circular representation of the plasmids of G. tundricola MP5ACTX9T displaying relevant genome features. From outside to center; Genes on forward strand (color by COG categories), genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content and GC skew. Order and size from left to right: pACIX901, 0.48 Mbp; pACIX902, 0.3 Mbp; pACIX903, 0.19 Mbp; pACIX904, 0.12 Mbp; pACIX905, 0.12 Mbp.
Discussion
Granulicella tundricola MP5ACTX9T is a tundra soil strain with a genome consisting of a circular chromosome and five mega plasmids ranging in size from 1.1 × 105 to 4.7 × 105 bp for a total genome size of 5.5 Mbp. The G. tundricola genome also contains close to twice as many pseudogenes and a large number of mobile genetic elements as compared to Granulicella mallensis and Terrigobus saanensis, two other Acidobacteria isolated from the same habitat [29]. A large number of genes assigned to COG functional categories for transport and metabolism of carbohydrates (6.9%) and amino acids (6.5%) and involved in cell envelope biogenesis (8%) and transcription (6.9%) were identified. Further genome analysis revealed an abundance of gene modules encoding for functional activities within the carbohydrate-active enzymes (CAZy) families [63,64] involved in breakdown, utilization and biosynthesis of carbohydrates. G. tundricola hydrolyzed complex carbon polymers, including CMC, pectin, lichenin, laminarin and starch, and utilized sugars such as cellobiose, D-mannose, D-xylose and D-trehalose. Genome predictions for CDSs encoding for enzymes such as cellulases, pectinases, alginate lyases, trehalase and amylases are in agreement with biochemical activities in strain MP5ACTX9T. However, the genome of G. tundricola did contain many CDSs encoding for GH18 chitinases although no chitinase activity was detected after 10 day-incubation with chitinazure [29]. In addition, the G. tundricola genome contained a cluster of genes in close proximity to the cellulose synthase gene (bcsAB), which included cellulase (bscZ) (endoglucanase Y) of family GH8, cellulose synthase operon protein (bcsC) and a cellulose synthase operon protein (yhjQ) involved in cellulose biosynthesis. We previously reported on a detailed comparative genome analysis of G. tundricola MP5ACTX9T with other Acidobacteria strains for which finished genomes are available [29]. The data suggests that G. tundricola is involved in hydrolysis and utilization of stored carbohydrates and biosynthesis of exopolysaccharides from organic matter and plant based polymers in the soil. Therefore, G. tundricola may be central to carbon cycling processes in Arctic and boreal soil ecosystems.
References
Männistö MK, Rawat S, Starovoytov V, Häggblom MM. Granulicella arctica sp. nov., Granulicella mallensis sp. nov., Granulicella sapmiensis sp. nov. and Granulicella tundricola sp. nov., novel Acidobacteria from tundra soil of Northern Finland. Int J Syst Evol Microbiol 2012; 62:2097–2106. PubMed http://dx.doi.org/10.1099/ijs.0.031864-0
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 2009; 3:442–453. PubMed http://dx.doi.org/10.1038/ismej.2008.127
Barns SM, Cain EC, Sommerville L, Kuske CR. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl Environ Microbiol 2007; 73:3113–3116. PubMed http://dx.doi.org/10.1128/AEM.02012-06
Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 2006; 72:1719–1728. PubMed http://dx.doi.org/10.1128/AEM.72.3.1719-1728.2006
Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology 2007; 88:1354–1364. PubMed http://dx.doi.org/10.1890/05-1839
Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EA. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol 2010; 12:1842–1854. PubMed http://dx.doi.org/10.1111/j.1462-2920.2010.02189.x
Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 2010; 12:2998–3006. PubMed http://dx.doi.org/10.1111/j.1462-2920.2010.02277.x
Pankratov TA, Dedysh SN. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer degrading acidobacteria from Sphagnum peat bogs. Int J Syst Evol Microbiol 2010; 60:2951–2959. PubMed http://dx.doi.org/10.1099/ijs.0.021824-0
Kishimoto N, Kosako Y, Tano T. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr Microbiol 1991; 22:1–7. http://dx.doi.org/10.1007/BF02106205
Eichorst SA, Breznak JA, Schmidt TM. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl Environ Microbiol 2007; 73:2708–2717. PubMed http://dx.doi.org/10.1128/AEM.02140-06
Männistö MK, Rawat SR, Starovoytov V, Häggblom MM. Terriglobus saanensis sp. nov., an acidobacterium isolated from tundra soil. Int J Syst Evol Microbiol 2011; 61:1823–1828. PubMed http://dx.doi.org/10.1099/ijs.0.026005-0
Koch IH, Gich F, Dunfield PF, Overmann J. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int J Syst Evol Microbiol 2008; 58:1114–1122. PubMed http://dx.doi.org/10.1099/ijs.0.65303-0
Okamura K, Kawai A, Yamada T, Hiraishi A. Acidipila rosea gen. nov., sp. nov., an acidophilic chemoorganotrophic bacterium belonging to the phylum Acidobacteria. FEMS Microbiol Lett 2011; 317:138–142. PubMed http://dx.doi.org/10.1111/j.1574-6968.2011.02224.x
Pankratov TA, Kirsanova LA, Kaparullina EN, Kevbrin VV, Dedysh SN. Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria and emended description of Acidobacterium capsulatum Kishimoto et al. Int J Syst Evol Microbiol 2012; 62:430–437. PubMed http://dx.doi.org/10.1099/ijs.0.029629-0
Kulichevskaya IS, Kostina LA, Valásková V, Rijpstra IC, Sinninghe Damsté JS, de Boer W, Dedysh SN. Acidicapsa borealis gen. nov., sp. nov. and A. ligni sp. nov., two novel subdivision 1 Acidobacteria from sphagnum peat and decaying wood. Int J Syst Evol Microbiol 2012; 62:1512–1520. PubMed http://dx.doi.org/10.1099/ijs.0.034819-0
Dedysh SN, Kulichevskaya IS, Serkebaeva YM, Mityaeva MA, Sorokin VV, Suzina NE, Rijpstra WI, Damste JS. Bryocella elongata gen. nov., sp. nov., a novel member of Subdivision 1 of the Acidobacteria isolated from a methanotrophic enrichment culture, and emended description of Edaphobacter aggregans Koch et al. 2008. Int J Syst Evol Microbiol 2012; 62:654–664. PubMed http://dx.doi.org/10.1099/ijs.0.031898-0
Kulichevskaya IS, Suzina NE, Liesack W, Dedysh SN. Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemoorganotroph from subdivision 3 of the Acidobacteria. Int J Syst Evol Microbiol 2010; 60:301–306. PubMed http://dx.doi.org/10.1099/ijs.0.013250-0
Foesel BU, Rohde M, Overmann J. Blastocatella fastidiosa gen. nov., sp. nov., isolated from semiarid savanna soil — The first described species of Acidobacteria subdivision 4. Syst Appl Microbiol 2013; 36:82–89. PubMed http://dx.doi.org/10.1016/j.syapm.2012.11.002
Izumi H, Nunoura T, Miyazaki M, Mino S, Toki T, Takai K, Sako Y, Sawabe T, Nakagawa S. Thermotomaculum hydrothermale gen. nov., sp. nov., a novel heterotrophic thermophile within the phylum Acidobacteria from a deep-sea hydrothermal vent chimney in the Southern Okinawa Trough. Extremophiles 2012; 16:245–253. PubMed http://dx.doi.org/10.1007/s00792-011-0425-9
Liesack W, Bak F, Kreft JU, Stackebrandt E. Holophaga foetida gen.nov., sp. nov., a new homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol 1994; 162:85–90. PubMed http://dx.doi.org/10.1007/BF00264378
Coates JD, Ellis DJ, Gaw CV, Lovley DR. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon contaminated aquifer. Int J Syst Bacteriol 1999; 49:1615–1622. PubMed http://dx.doi.org/10.1099/00207713-49-4-1615
Fukunaga Y, Kurahashi M, Yanagi K, Yokota A, Harayama S. Acanthopleuribacter pedis gen. nov., sp. nov., a marine bacterium isolated from a chiton, and description of Acanthopleuribacteraceae fam. nov., Acanthopleuribacterales ord. nov., Holophagales ord. nov. and Holophagae classis nov. in the phylum ‘Acidobacteria’. Int J Syst Evol Microbiol 2008; 58:2597–2601. PubMed http://dx.doi.org/10.1099/ijs.0.65589-0
Losey NA, Stevenson BS, Busse HJ, Damste JSS, Rijpstra WIC, Rudd S, Lawson PA. Thermoanaerobaculum aquaticum gen. nov., sp. nov., the first cultivated member of Acidobacteria subdivision 23, isolated from a hot spring. [PMID: 23771620]. [DOI 10.1099/ijs.0.051425-0]. Int J Syst Evol Microbiol 2013; 63:4149–4157.
Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 2009; 75:2046–2056. PubMed http://dx.doi.org/10.1128/AEM.02294-08
Bryant DA, Amaya M. Garcia Costas AMG, Maresca JA, Chew AGM, Klatt CG, Bateson MM, Tallon LJ, Hostetler J, Nelson WC, Heidelberg JF, Ward DM. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 2007; 317:523–526. PubMed http://dx.doi.org/10.1126/science.1143236
Männistö MK, Tiirola M, Häggblom MM. Microbial communities in Arctic fjelds of Finnish Lapland are stable but highly pH dependent. FEMS Microbiol Ecol 2007; 59:452–465. PubMed http://dx.doi.org/10.1111/j.1574-6941.2006.00232.x
Sait M, Davis KE, Janssen PH. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl Environ Microbiol 2006; 72:1852–1857. PubMed http://dx.doi.org/10.1128/AEM.72.3.1852-1857.2006
Eichorst SA, Kuske CR, Schmidt TM. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl Environ Microbiol 2011; 77:586–596. PubMed http://dx.doi.org/10.1128/AEM.01080-10
Rawat SR, Männistö MK, Bromberg Y, Häggblom MM. Comparative genomic and physiological analysis provides insights into the role of Acidobacteria in organic carbon utilization in Arctic tundra soils. FEMS Microbiol Ecol 2012; 82:341–355. PubMed http://dx.doi.org/10.1111/j.1574-6941.2012.01381.x
Rawat SR, Männistö MK, Starovoytov V, Goodwin L, Nolan M, Hauser L, Land M, Davenport KW, Woyke T, Häggblom MM. Complete genome sequence of Terriglobus saanensis strain SP1 PR4T, an Acidobacteria from tundra soil. Stand Genomic Sci 2012; 7:59–69. PubMed http://dx.doi.org/10.4056/sigs.3036810
Männistö MK, Tiirola M, Häggblom MM. Effect of freeze-thaw cycles on bacterial communities of Arctic tundra soil. Microb Ecol 2009; 58:621–631. PubMed http://dx.doi.org/10.1007/s00248-009-9516-x
Männistö MK, Kurhela E, Tiirola M, Häggblom MM. Acidobacteria dominate the active bacterial communities of sub-Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol Ecol 2013; 84:47–59. PubMed http://dx.doi.org/10.1111/1574-6941.12035
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed http://dx.doi.org/10.1073/pnas.87.12.4576
Thrash JC, Coates JD. Phylum XVII. Acidobacteria phyl. nov. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 4, Springer, New York, 2011, p. 725.
Validation List No. 143. Int J Syst Evol Microbiol 2012; 62:1–4. http://dx.doi.org/10.1099/ijs.0.039487-0
Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 2002; 52:7–76. PubMed
Judicial Commission of the International Committee on Systematics of Prokaryotes. The nomenclatural types of the orders Acholeplasmatales, Halanaerobiales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales, Planctomycetales, Prochlorales, Sulfolobales, Thermococcales, Thermoproteales and Verrucomicrobiales are the genera Acholeplasma, Halanaerobium, Halobacterium, Methanobacterium, Methanococcus, Methanomicrobium, Planctomyces, Prochloron, Sulfolobus, Thermococcus, Thermoproteus and Verrucomicrobium, respectively. Opinion 79. Int J Syst Evol Microbiol 2005; 55:517–518. PubMed http://dx.doi.org/10.1099/ijs.0.63548-0
Ludwig W, Euzeby J, Whitman WG. Draft taxonomic outline of the Bacteroidetes, Planctomycetes, Chlamydiae, Spirochaetes, Fibrobacteres, Fusobacteria, Acidobacteria, Verrucomicrobia, Dictyoglomi, and Gemmatimonadetes. http://www.bergeys.org/outlines/Bergeys_Vol_4_Outline.pdf. Taxonomic Outline 2008.
Thrash JC, Coates JD. Family I. Acidobacteriaceae fam. nov. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 4, Springer, New York, 2011, p. 728.
Pankratov TA, Dedysh SN. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int J Syst Evol Microbiol 2010; 60:2951–2959. PubMed http://dx.doi.org/10.1099/ijs.0.021824-0
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. PubMed http://dx.doi.org/10.1038/75556
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–2739. PubMed http://dx.doi.org/10.1093/molbev/msr121
Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2007; 36:D475–D479. PubMed http://dx.doi.org/10.1093/nar/gkm884
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed http://dx.doi.org/10.1038/nbt1360
DOE Joint Genome Institute. http://www.jgi.doe.gov.
Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433–438. PubMed http://dx.doi.org/10.1517/14622416.5.4.433
Margulies M, Egholm M, Altman WE. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:376–380. PubMed
Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821–829. PubMed http://dx.doi.org/10.1101/gr.074492.107
Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175–185. PubMed http://dx.doi.org/10.1101/gr.8.3.175
Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195–202. PubMed http://dx.doi.org/10.1101/gr.8.3.195
The Phred/Phrap/Consed software package. http://www.phrap.com.
Han CS, Chain P. Finishing repeat regions automatically with Dupfinisher CSREA Press. In: Arabnia AR, Valafar H, editors. Proceedings of the 2006 international conference on bioinformatics & computational biology; 2006; June 26–29. CSREA Press. p 141–146.
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119. PubMed http://dx.doi.org/10.1186/1471-2105-11-119
Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455–457. PubMed http://dx.doi.org/10.1038/nmeth.1457
Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science 1997; 278:631–637. PubMed http://dx.doi.org/10.1126/science.278.5338.631
Clusters of Orthologous Groups. http://www.ncbi.nlm.nih.gov/COG.
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955–964. PubMed
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100–3108. PubMed http://dx.doi.org/10.1093/nar/gkm160
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res 2003; 31:439–441. PubMed http://dx.doi.org/10.1093/nar/gkg006
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Bol 2001; 305:567–580. PubMed http://dx.doi.org/10.1006/jmbi.2000.4315
Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783–795. PubMed http://dx.doi.org/10.1016/j.jmb.2004.05.028
Markowitz VM, Mavromatis K, Ivanova N, Chen IM, Chu K, Kyrpides N. Expert Review of Functional Annotations for Microbial Genomes. Bioinformatics 2009; 25:2271–2278. PubMed http://dx.doi.org/10.1093/bioinformatics/btp393
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 2009; 37:D233–D238. PubMed http://dx.doi.org/10.1093/nar/gkn663
Lombard V, Ramulu HG, Drula E, Coutinho PM and Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research 1–6.
Acknowledgements
The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy Under Contract No. DE-AC02-05CH11231. This work was funded in part by the Academy of Finland and the New Jersey Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Rawat, S.R., Männistö, M.K., Starovoytov, V. et al. Complete genome sequence of Granulicella tundricola type strain MP5ACTX9T, an Acidobacteria from tundra soil. Stand in Genomic Sci 9, 449–461 (2014). https://doi.org/10.4056/sigs.4648353
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.4648353