Abstract
Background
Myxobacteria harbor numerous biosynthetic gene clusters that can produce a diverse range of secondary metabolites. Minicystis rosea DSM 24000T is a soil-dwelling myxobacterium belonging to the suborderSorangiineae and family Polyangiaceae and is known to produce various secondary metabolites as well as polyunsaturated fatty acids (PUFAs). Here, we use whole-genome sequencing to explore the diversity of biosynthetic gene clusters in M. rosea.
Results
Using PacBio sequencing technology, we assembled the 16.04 Mbp complete genome of M. rosea DSM 24000T, the largest bacterial genome sequenced to date. About 44% of its coding potential represents paralogous genes predominantly associated with signal transduction, transcriptional regulation, and protein folding. These genes are involved in various essential functions such as cellular organization, diverse niche adaptation, and bacterial cooperation, and enable social behavior like gliding motility, sporulation, and predation, typical of myxobacteria. A profusion of eukaryotic-like kinases (353) and an elevated ratio of phosphatases (8.2/1) in M. rosea as compared to other myxobacteria suggest gene duplication as one of the primary modes of genome expansion. About 7.7% of the genes are involved in the biosynthesis of a diverse array of secondary metabolites such as polyketides, terpenes, and bacteriocins. Phylogeny of the genes involved in PUFA biosynthesis (pfa) together with the conserved synteny of the complete pfa gene cluster suggests acquisition via horizontal gene transfer from Actinobacteria.
Conclusion
Overall, this study describes the complete genome sequence of M. rosea, comparative genomic analysis to explore the putative reasons for its large genome size, and explores the secondary metabolite potential, including the biosynthesis of polyunsaturated fatty acids.
Similar content being viewed by others
Background
Myxobacteria are Gram-negative, rod-shaped, soil-dwelling δ-proteobacteria taxonomically classified within the order Myxococcales and distributed across diverse ecological niches [1,2,3]. While the δ-proteobacteria are anaerobic sulfate or sulfur-reducing microbes, myxobacteria are aerobes except for the facultative anaerobe Anaeromyxobacter spp. and the strictly anaerobic Pajaroellobacter spp. [4, 5]. Unlike their close δ-proteobacteria relatives, they have large genomes (9–16 Mbp) with the exception of Anaeromyxobacter spp. (~ 5 Mbp), Vulgatibacter (4.35 Mbp), and Pajaroellobacter (1.82 Mbp). Apart from cellular functions, most of the functionally annotated proteins are associated with several intriguing physiological characteristics such as gliding motility, predation, fruiting body formation, biofilm formation, social behavior, etc. [6,7,8,9,10,11,12,13]. Myxobacterial vegetative cells can swarm by social and adventurous gliding in search of nutrients or for predating other microbes [3]. During starvation, myxobacterial cells (>105) construct fruiting bodies which enclose myxospores that can initiate their vegetative cycle in favorable growth conditions [14].
Myxobacteria are known for their vast biosynthetic potential, as evident by the secretion of a large variety of bioactive molecules such as alkaloid, polyketide, terpene, aminocoumarin, beta-lactam, etc., produced from polyketide synthase (PKS), nonribosomal polypeptide synthetase (NRPS), and their hybrids [15, 16]. These compounds are known to have various antibiotic, antifungal and antitumor activities [17]. Most of these studied organisms belonging to Sorangium and Aetherobacter have been reported as potent producers of polyunsaturated fatty acids (PUFAs), including eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) [18]. These n-3 (omega-3) and n-6 (omega-6) are associated with blood-pressure-lowering properties and are used for the treatment of cardiovascular diseases, diabetes, and obesity [19]. Fish oils are well-known eukaryotic sources of DHA and EPA [20] but might be contaminated with organic pollutants. Considering the huge demand for PUFA due to its health benefits, alternate PUFA synthesis via an anaerobic route integrated with a polyketide synthase (PKS) instead of the fatty acid synthase (FAS) has been explored in prokaryotes [21, 22]. This pathway for PUFA synthesis employs pfa gene clusters containing a total of five consecutive genes (pfaA, pfaB, pfaC, pfaD, and pfaE) in marine microorganisms such as S. pneumatophori SCRC-2738, M. marina MP-1, and P. profundum SS9 [22,23,24]. Recently, these pfa gene clusters have also been explored in non-marine terrestrial myxobacteria Aetherobacter sp. SBSr008, Aetherobacter fasciculatus SBSr002, and S. cellulosum So ce56 in producing arachidonic acid (ARA), DHA, EPA as well as linolenic acid (LA), γ-linolenic acid (GLA), stearidonic acid (SDA), and docosapentaenoic acid (DPA) [18]. Unlike marine microorganisms, myxobacterial pfa gene clusters include only four genes i.e., pfa1 (homolog of pfaD), pfa2 (homolog of pfaA), pfa3 (homolog of pfaC), and a homolog of pfaE gene [18]. These Pfa proteins contain various domains and catalytic sites such as Pfa1 (PfaD) in Aetherobacter contains enoyl reductase (ER) domain, multi-functional Pfa2 (PfaA) protein contains several domains, i.e. β-ketoacyl synthase (KS), malonyl/acyltransferase (MAT/AT), acyl carrier protein (ACP), ketoreductase (KR) and PKS-like dehydratase (PS-DH) domains; and Pfa3 (PfaC) has KS, chain length factor (CLF), acyltransferase (AT), Fab-A like dehydratase (DH), pseudo-domain dehydratase (DH’) and 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) domain. In addition to these consecutive genes, another gene pfaE encodes for 4′-phosphopantetheinyl transferase (PPTase) [25] and is located at a separate locus in Aetherobacter and S. cellulosum So ce56 genomes. These domains are not similarly distributed in myxobacterial proteomes [18]. For example, the AT domain, seen in Pfa3 of Aetherobacter, is not present in S. cellulosum So ce56. Diversity of these domains have been reported to cause product variations from pfa gene clusters of terrestrial myxobacteria [18].
To characterize and explore the huge biosynthetic potential of myxobacteria, whole-genome sequencing of more strains is needed. Here we report the complete genome sequence of M. rosea DSM 24000T and identify several biosynthetic gene clusters including one involved in the synthesis of PUFA. We also perform comparative genome analysis of M. rosea and other related myxobacteria to glean insights about the expansion in genome size that makes the M. rosea DSM 24000T genome the largest bacterial genome known to date.
Results and discussion
Genomic properties of M. rosea DSM 24000T
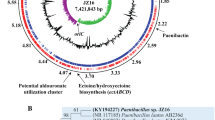
The M. rosea genome assembled into a complete circular chromosome of length 16,040,666 bp with 69.07% GC. It has been deposited in GenBank under the accession number CP016211.1 within the BioProject number PRJNA321464. The assembly process did not detect any plasmid sequence. This is not surprising as among the genomes of order Myxococcales, only one organism M. fulvus 124B02 has been reported to harbor a plasmid, pMF1 [26]. However, as we have used Bluepippin size selection in our sequencing, we might have missed any smaller size plasmid. RAST-based annotation has predicted 14,121 genes that consist of 14,018 protein-coding genes, 88 tRNAs, four 5S–16S-23S rRNA operons, one transfer-messenger RNA, and two non-coding RNAs (each belongs to RNase_P_RNA and SRP_RNA class) (Table 1). As of date, the genome sequence of M. rosea is the largest amongst kingdom bacteria (Fig. 1), and is ~ 1.26 Mbp larger than the genome of the myxobacteria S. cellulosum So0157–2 (14,782,125 bp), which has been previously reported as the largest prokaryotic genome [27].
Circular representation of the genome of M. rosea DSM 24000T showing GC skew, GC content, genes on leading and lagging strands, core genes, duplicate genes, unique genes, unique exogenous genes, secondary metabolites producing genes (BGCs), and eukaryotic-like kinase (ELK) synthesizing genes from inner to outer layers respectively
16S rRNA-based phylogenetic tree indicates that M. rosea DSM 24000T is a close relative of members of the family Polyangiaceae in suborder Sorangiineae (Fig. 2). Similar tree topology has also been observed in the marker-gene-based tree where M. rosea is closely clustered with selected species of the genera within the Polyangiaceae family (Fig. S1). Moreover, M. rosea also shows higher DDH and ANI values with the Sorangium spp. as compared to other myxobacteria (Table S1) suggesting their close relatedness.
Analysis of genome expansion and protein function in M. rosea DSM 24000T
M. rosea encodes 14,018 protein-coding sequences which account for 87.50% coding density with an average gene size of 1003 bp (Table 1). A total of 6,167 (~ 44%) coding sequences have been annotated as hypothetical proteins in M. rosea. Our pan-genome studies with 19 other myxobacteria (having > 9 Mbp genome size) revealed vast diversity among all studied members.
Core genome
Our study suggested that 650 orthologous protein-coding genes are conserved and constitute the core genome. This category includes only 5.03% of M. rosea proteins in contrast with its vast gene content (Table S2a). COG-based functional characterization of core proteins in M. rosea reveals that ‘Metabolism’ [MET] (44.14%) representation is higher than ‘Information Storage and Processing’ [ISP] (28.76%) and ‘Cellular Processes and Signaling’ [CPS] (27.70%). Most of the core proteins in M. rosea are involved in translation [J] (16.59%), coenzyme metabolism [H] (8.68%), lipid metabolism [I] (8.07%), energy production [C] (7%), post-translational modification [O] (6.85%), amino acid transport [E] (6.39%), transcription [K] (6.24%), cell wall biogenesis [M] (5.78%), replication [L] (5.78%), nucleotide metabolism [F] (5.33%), and signal transduction [T] (5.02%) (Fig. S2).
Accessory genome
This study identified a total of 8947 (63.83%) accessory genes in M. rosea (Fig. 3), which are associated with the COG category CPS in higher number (39.29%) as compared to the MET (36.86%) and ISP (16.74%) categories. Most of the accessory proteins are involved in signal transduction [T] (17.02%), transcription [K] (10.57%), cell wall biogenesis [M] (7.33%), lipid metabolism [I] (6.56%), amino acid transport [E] (5.67%), energy production [C] (5.33%), and secondary metabolites biosynthesis [Q] (5.19%) (Fig. S2).
Unique genome
A total of 4421 (31.54%) proteins do not display any significant identity with selected myxobacteria, which are mentioned as unique proteins in M. rosea (Table S2a). Among them, only 347 unique proteins have been functionally identified which are associated with the COG category CPS (34%) followed by MET (30.25%) and ISP (13.83%). Majority of unique known proteins are involved in signal transduction [T] (12.68%), transcription [K] (10.29%), cell wall biogenesis [M] (9.80%), lipid metabolism [I] (5.76%), secondary metabolites biosynthesis [Q] (5.19%), coenzyme metabolism [H] (4.61%), and post-translational modification [O] (4.32%) (Fig. S2). Among unique proteins in M. rosea, 125 proteins exhibit significant similarity with exogenous genetic materials, including integrated plasmids, phages, and insertion sequence (IS) elements (Table S2a). Twenty-four genomic islands (GIs) have been identified in M. rosea comprising a total of 6,15,248 bp (3.84%) of the genome (Table S2b). The GIs containing unique exogenous genes (Table S2b) may help facilitate horizontal gene transfer [28].
Signal transduction
Overall, our genome analysis indicates an abundance of signal transduction proteins as well as transcriptional regulators in M. rosea. Our analysis is supported by previous studies reporting a strong correlation between the number of bacterial transcriptional regulators and genome size [29]. Earlier, a linear relationship has been observed between the signaling proteins, including two-component system (TCS) proteins, and genome size in host-associated, as well as, environmental bacteria [30]. M. rosea also shows a higher number (323 proteins) of TCS proteins, which comprise 145 orphan histidine kinases (HK), 125 orphan response regulators (RR), and 53 hybrid TCS proteins as compared to S. cellulosum So0157–2 (309 TCS proteins) as well as other Sorangium spp. (Fig. 4A). However, no strong correlation (r = 0.531, p < 0.05) between the genome size and the number of TCS proteins has been found in myxobacteria, as reported previously [9]. Apart from the environmental diversity, the complex life cycle also influences the numbers of TCS proteins in the case of myxobacteria [31]. In addition to the TCS system, signal transduction mechanisms are also facilitated by serine, threonine, and tyrosine phosphorylation mediated protein kinases in prokaryotes. This protein family in myxobacteria has been reported to have strong sequence similarity with eukaryotic-like kinases (ELKs) [32]. M. rosea contains 353 ELKs, which is higher than S. cellulosum So ce56 (317) [33], as well as other myxobacteria (Fig. 4B). The number of ELKs increases with increasing genome size in bacteria [34]. A significant strong positive correlation between genome size and number of ELKs (r = 0.859, p < 0.001) is seen. In contrast to ELKs, M. rosea has fewer protein phosphatases (PPs) (43 genes), comprising all three major families of PPs i.e., serine/threonine PPs (PPP-family = 9 genes), metal-dependent serine/threonine PPs (PPM-family) including PP2c-type (21 genes) and SpoIIE-like PPs (5 genes), and tyrosine-specific PPs (PTP-family) including dual-specificity PTPs (5 genes), low molecular weight protein PTPs (2 genes) and PTPZ-like PTPs (1 gene). In response to the peripheral stimuli, protein kinases phosphorylate the target proteins, whereas, phosphatases deactivate them by removing the phosphate groups [35]. Thus, kinase/phosphatase ratio regulates the bacterial cell differentiation and development to quickly adapt to the persistently varying environment [36]. It has also been reported that PP2c-type PPs can compete with ELKs in bacteria [37]. However, a higher number of PP2c-type PPs has been observed in M. rosea (21 genes) than A. dehalogenans (2 genes), M. xanthus (4 genes), and S. cellulosum So ce56 (16 genes), reported as the highest PP2c-type PPs containing prokaryote [38]. Moreover, an elevated ratio of ELKs/PPs has been also observed in M. rosea (8.2/1), as in A. dehalogenans (1.7/1), M. xanthus (6.9/1), and S. cellulosum So ce56 (7.7/1) [38]. It could explain the phosphorylation events which cannot be reversed by PPs during multicellular development in myxobacteria [38]. We identified 90 ELK proteins as being involved in the fruiting body production in M. rosea by BLASTP search (length ≥ 50% and e-value ≤1e-10) against the fruiting body forming proteins of M. xanthus [39] and HMM-profile based searches [40]. However, crucial genes for fruiting body development (actA, asgA, csgA, fruA, and sdeK) identified in M. xanthus are absent in M. rosea and in S. cellulosum So ce56 [33]. Therefore, as suggested in earlier studies [41], it can be argued that an alternative mechanism for fruiting body development may exist in M. rosea [42].
Secretome analysis
Our analysis revealed that 3035 proteins constitute the secretome in M. rosea, which is higher as compared to other myxobacteria (Fig. 4C). Significant positive correlation is seen between genome size and the number of secretome proteins (r = 0.845, p < 0.001). KEGG pathway analysis [43] has also revealed a higher number of proteins (104 proteins) are involved in the secretion system in M. rosea (KEGG pathway ID - mrm03070) as compared to A. dehalogenans 2CP-1 (47 proteins), C. fuscus DSM 2262 (60 proteins), A. gephyra DSM 2261T (58 proteins), M. hansupus (53 proteins), L. luteola DSM 27648T (35 proteins), S. amylolyticus DSM 53668T (44 proteins), S. cellulosum So ce56 (67 proteins), S. cellulosum So0157–2 (64 proteins), and V. incomptus DSM 27710T (27 proteins). An extensive secretion system may explain the selection of such a large number of associated genes in M. rosea for executing sophisticated cellular crosstalk and adaptation to diverse environments.
A variety of regulatory systems are broadly distributed across the M. rosea proteome, with most of them involved in transcription regulation. Free-living and soil-dwelling large-genome-containing bacteria usually acquire a complex regulatory network and a higher number of corresponding genes to survive in environments where the resources for growth are scarce but diverse [44]. Moreover, a higher number of lipid metabolism [I] associated proteins than carbohydrate metabolism [G] reveals efficient utilization of lipid as an energy source in M. rosea similar to that observed in M. xanthus [45]. Lipids have been observed in producing diverse morphological characters such as fruiting body formation in myxobacteria upon amino acid and carbon depletion [46]. Steroid biosynthesis in M. rosea further explores the importance of lipid bodies as signaling molecules similar to the steroid hormones in animals [47]. Thus, sophisticated intercellular communication for niche adaptation and morphogenetic variations may facilitate the retention of a huge amount of protein-coding genes in M. rosea.
Duplication events
Paralogous genes, which arise by gene duplications, comprise 44.10% genes in M. rosea (Table S2a). Using the same parameters to define paralogous genes, we find that well-studied members of the family Polyangiaceae i.e., S. cellulosum So ce56 and S. cellulosum So0157–2 contain 47.10 and 41.80% paralogous genes, respectively. Our results are in agreement with previous reports suggesting that the extensive expansion of paralogous genes account for the large genome size [48], similar to that reported in S. cellulosum So ce56 [33] and S. cellulosum So0157–2 [27]. Most of the functionally annotated paralogous proteins are involved in signal transduction [T] (21.41%), transcription [K] (12.04%), cell wall biogenesis [M] (8.08%), lipid metabolism [I] (7.30%), post-translational modification [O] (6.89%), and biosynthesis of secondary metabolites [Q] (5.47%) in M. rosea. Thus, the majority of gene duplications have occurred for those proteins in M. rosea that may help it to respond to the environmental signals and in regulatory mechanisms for niche adaptation.
Pfam-based functional characterization
Using HMM profile-based searches, we identified that 7446 M. rosea proteins were mapped to 2576 Pfam families. Comparative analysis of protein families reveals that several families such as protein kinase (360 members); histidine kinase (344 members); helix-turn-helix (315 members), TetR (139 members), transcription regulators like σ54 (104 members); repeats such as tetratricopeptide repeats (134 members), pentapeptide repeats (107 members), VCBS (91 members), Sel1 (16 members); phage_GPD (71 members); FGE-sulfatase (69 members); short-chain dehydrogenase (115 members); and radical SAM (70 members) are overrepresented in M. rosea as compared to other Sorangiineae members (C. apiculatus, L. luteola, Polyangium spp., S. amylolyticus, and S. cellulosum) (Table S2c). These families are associated with signaling systems, regulatory networks, protein folding, and genome packaging in M. rosea. Apart from these, some families such as, abhydrolase_7, aerolysin, bile_hydr_trans, creD, disintegrin, endonuclease_1, endotoxin_N, expansin_C, gluconate_2-dh3, gly_transf_sug, glyco_hydro, lectin_legB, lipase_bact_N, lipocalin, peptidase_C2, TPP_enzyme_M_2, etc. are exclusively identified in M. rosea (Table S2c). Complex lifestyles in diverse environments might facilitate gene gain, loss, or duplication in microbes for adaptation to that niche [49]. The retention/modification of duplicated genes helps to conserve the protein functions amongst different environments [50], which could be one of the predominant causes for the large genome size in M. rosea.
Biosynthetic gene clusters in M. rosea DSM 24000T especially polyunsaturated fatty acid (PUFA) biosynthetic genes
Genome mining revealed 47 BGCs (encoded by 1081 genes) comprising 7.71% of protein-coding genes in M. rosea. The major fraction of biosynthetic genes encode NRPS (252 genes; 7 clusters) followed by terpene (171 genes; 9 clusters), PKS (128 genes; 4 clusters), ribosomal synthesized and post-translationally modified peptide (RiPP) (75 genes; 7 clusters), arylpolyene (75 genes; 2 clusters), lanthipeptide (73 genes; 3 clusters), RRE-containing (70 genes; 4 clusters), indole (56 genes; 3 clusters), and NRPS-PKS hybrid (30 genes; 1 cluster). Other clusters such as phosphonate (1 cluster; 38 genes), thioamitide (2 clusters; 32 genes), thiopeptide (1 cluster; 30 genes), phenazine (1 cluster; 20 genes), LAP (1 cluster; 18 genes), and siderophore (1 cluster; 13 genes) are also detected in the M. rosea genome. The representation of BGC genes in the M. rosea genome is more than the average bacterial genome (3.7%) and is similar to organisms from the genus Streptomyces, Myxococcus, Sorangium, and Burkholderia [51].
Further analysis of PKSs in M. rosea DSM 24000T reveals the pfa gene cluster comprises four genes (pfa1, pfa2, pfa3, and pfaE) (Fig. 5) as observed in Aetherobacter and Sorangium [18]. Domain analysis of the respective proteins shows that Pfa1 (PfaD) [A7982_11504] contains a nitronate monooxygenase domain of enoyl reductase (ER) (Fig. 5AI). Sequence similarity and domain conservation of Pfa1 are seen between M. rosea and Aetherobacter spp. (Fig. 5BI). Several functional domains, i.e., β-ketoacyl synthase (KS), malonyl/acyltransferase (MAT/AT), acyl carrier protein (ACP), ketoreductase (KR), and PKS-like dehydratase (PS-DH) are positioned similarly in Pfa2 of M. rosea [A7982_11505] and Aetherobacter spp. (Fig. 5AI). The Pfa2 protein-based phylogenetic tree also reveals close relatedness between Aetherobacter spp. and M. rosea (Fig. 5BII). Pfa3 in M. rosea [A7982_11506] comprises several domains such as KS, chain length factor (CLF), acyltransferase (AT), Fab-A-like dehydratase (DH), pseudo-domain dehydratase (DH’), and 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) (Fig. 5AI) as detected in Aetherobacter spp. (Fig. 5AII). A close phylogenetic relationship is also present between the Pfa3 protein of M. rosea and Aetherobacter spp. (Fig. 5BIII). The KS domain catalyzes condensation reaction for fatty acid chain elongation [52], whereas CLF controls the fatty acid chain length [53]. MAT/AT acts as a chain extender by selecting and transferring malonic esters with the help of ACP. Other enzymes like KR, DH, and ER introduce structural diversity in the fatty acid chain. They act as tailoring enzymes that reduce intermediate keto groups, thus modifying the nascent fatty acid chain [54]. The integration of AGPAT domain into Pfa3 protein has been reported as a unique feature of the terrestrial myxobacterial PUFA synthases, which catalyzes acyl group’s transfer to generate phosphatidic acid in the chain-terminating step of PUFA synthesis [55]. Posttranslational modification of ACP occurs by the phosphopantetheinylation that converts apo-ACP to an active holo form by 4′-phosphopantetheinyl transferase (PPTase) [25]. PPTase domain has been observed in PfaE protein [A7982_13498] which is located at a separate locus of M. rosea proteome (Fig. 5AI) as observed in other myxobacteria like Aetherobacter (Fig. 5AII) and Sorangium (Fig. 5AIII) [18].
PUFA biosynthetic gene cluster organization, their phylogeny, and synteny analysis: A PUFA biosynthetic gene clusters and their respective domains in M. rosea DSM 24000T (I), Aetherobacter sp. SBSr008 (II), and S. cellulosum So ce56 (III). B Maximum likelihood-based phylogenetic analysis shows close relatedness of the M. rosea DSM 24000T PUFA biosynthetic proteins: Pfa1 [A7982_11504/APR86155.1] (I), Pfa2 [A7982_11505/APR86156.1] (II), and Pfa3 [A7982_11506/APR86157.1] (III) in members belonging to the genus Aetherobacter, Sorangium, Streptomyces, Azospirillum, Tahibacter, etc. C Synteny analysis of pfa gene cluster in M. rosea DSM 24000T with the close relatives belonging to the genus Aetherobacter, Sorangium, Streptomyces, Azospirillum, Tahibacter, etc.
The acyltransferase (AT) domain is distinctly encoded by PfaB in marine γ-proteobacteria such as S. pneumatophori SCRC-2738, P. profundum SS9 [22, 23], and M. marina MP-1 [24]. Whereas, AT domain is integrated into the carboxy-terminus of pfa3 in M. rosea (Fig. 5AI) as observed in terrestrial myxobacteria Aetherobacter (Fig. 5AII) [18]. The domain shows 65.26% and 64.91% identities with the AT domains of pfa3 proteins in Aetherobacter fasciculatus and Aetherobacter sp. SBSr008, respectively. It plays a significant role in shaping the final PUFA products synthesized from the PUFA gene cluster. However, the AT domain is not present in pfa3 of Sorangium (Fig. 5AIII), which has been suggested as the reason for the inability of Sorangium to produce DHA and EPA [18]. Overall, homology studies suggest that the PUFA clusters in M. rosea and Aetherobacter are unique amongst myxobacteria, containing all ten enzyme domains to yield PUFAs [56] including ARA, DHA, EPA as well as LA, GLA, SDA, and DPA. The fully functional PUFA synthase in M. rosea enables it to produce approximately 30% of the total cellular fatty acids [57]. Overall, the phylogeny of each gene (Fig. 5B) within the PUFA cluster reveal that these PUFA genes are evolutionarily closely related to Actinobacteria, suggesting that M. rosea might have acquired these genes from Streptomyces species via horizontal gene transfer.
To further confirm how this cluster evolved within M. rosea, we also performed synteny studies based on identified homologs across close relatives. We identified that the pfa gene cluster in M. rosea along with close relatives Aetherobacter and Sorangium is completely conserved with the PUFA synthetic gene cluster in several Streptomyces spp., Azospirillum melinis, Tahibacter aquaticus, etc. (Fig. 5C). Conclusively, based on our phylogenetic and synteny analysis, we speculate that the pfa gene cluster might have been horizontally transferred to M. rosea and closely related myxobacteria i.e., Aetherobacter and Sorangium from Actinobacteria.
Conclusions
Myxobacteria are well known for their large genome size and genomic content, as well as the potential to produce a wide range of secondary metabolites, including polyunsaturated fatty acids. Although there has been a huge surge in next-generation sequencing of microbes in the last three decades, however, in comparison to other soil bacteria, only a few whole-genome sequences of myxobacteria are available. In the present work, we have sequenced, assembled, and annotated a 16.04 Mbp circular genome of M. rosea DSM 24000T, the largest bacterial genome sequenced to date along with its genome characterization, and further emphasized the putative reasons for its genome expansion. Phylogenetic analysis and genome-genome distance calculation suggest M. rosea to be a close relative of the members of suborder Sorangiineae in the family Polyangiaceae. Due to its complex social behavior, diverse niche adaptation, and large genome size, M. rosea encodes a plethora of genes. Analysis of protein families reveals that most of the functionally identified proteins are associated with regulatory functions, protein folding, and genome packaging. Overrepresentation of protein families such as protein kinase, histidine kinase, tetR, transcription regulators like σ54, tetratricopeptide and pentapeptide repeats, VCBS, sel1, phage_GPD, FGE-sulfatase, short-chain dehydrogenase, and radical SAM, as well as higher numbers of secretomes and eukaryotic-like kinases in M. rosea as compared to other myxobacteria, are important explanations for genome expansion. Therefore, the requisite of adaptation in varied niches and complex myxobacterial multicellular behavior could be the driving forces behind genome expansion in M. rosea, which might be facilitated via gene-duplication followed by functional diversification of these proteins. A vast number of biosynthetic genes (7.71% of the coding potential) reveals the diversity of secondary metabolites production in M. rosea. Our study has identified the previously known functional PUFA biosynthetic gene cluster in the genome, one of the few known prokaryotic sources of DHA, EPA, LA, GLA, SDA, and DPA. Additionally, based on our phylogenetic and synteny studies, we hypothesize that this cluster might have been horizontally transferred from Actinobacteria. Our study on the genome sequencing, functional characterization, and pfa gene cluster analysis of M. rosea could further help biotechnological areas for heterologous expression of PUFAs from prokaryotes.
Materials and methods
Bacterial culture and isolation of genomic DNA
The actively growing plate culture of M. rosea was procured from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) as strain number DSM 24000T (also known as strain SBNa008 or NCCB 100349). The colonies from the procured sample were subcultured on VY/2 agar (DSMZ Medium 9) plates. These actively growing subculture plates were used to isolate whole genomic DNA using Zymogen Research Bacterial/fungal DNA isolation kit and Phenol-Chloroform-Isoamyl alcohol (PCI) methods. The quantity and quality of the extracted DNA were confirmed by gel electrophoresis and Nanodrop and supported by Qubit quantification.
Genome sequencing and assembly of M. rosea DSM 24000T
Isolated high-quality DNA was used for whole-genome sequencing (WGS) on a Pacific Biosciences RSII instrument available at the McGill University and Genome Quebec Innovation Center, Montreal (Quebec), Canada. SMRTbell long library was created with 10 mg whole genomic DNA using a 20-kb template preparation method using Procedure and Checklist-20 kb Template Preparation using BluePippin™ Size Selection (https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-gDNA-Libraries-Using-the-SMRTbell-Express-Template-Preparation-Kit-2.0.pdf; last accessed: 3 Sep 2021). Later the library was loaded onto three single molecules real-time (SMRT) cells and sequenced using P6 polymerase and C4 chemistry (P6C4) with 180-min movie time. PacBio sequencing generated 4,41,539 raw reads (3,48,84,02,643 bp) with an average read length of 7900 bp. The Hierarchical Genome Assembly Process (HGAP) Pipeline v. SMRT v2.3.0 and consensus polishing with Quiver [58] were used to generate de novo assembly using default parameters. Gene prediction and functional annotation were performed by Rapid Annotation using Subsystem Technology (RAST) [59], whereas rRNA and tRNA genes were predicted using RNAmmer 1.2 [60] and tRNAscan-SE-1.23 [61]. RNAz 2.0 tool [62] was used to identify structured non-coding RNA (P > 0.85). A circular plot for M. rosea DSM 24000T genome was drawn using BRIG (v 0.95-dev.0004) [63].
Phylogenetic analysis and estimation of DNA-DNA hybridization and average nucleotide identity
The 16S rRNA sequences reported for the members of all three myxobacteria suborders i.e., Cystobacterineae, Nannocystineae, and Sorangiineae were retrieved from the NCBI database. 16S rRNA sequences from all myxobacteria and an outgroup D. retbaense DSM 5692 were aligned using ClustalW [64]. The alignment was used to generate a phylogenetic tree using the GTR-GAMMA model [bootstrap: 100] of maximum likelihood (ML) method in the RAxML (v8) tool [65] and visualized by iTOL [66]. We also performed phylogenetic analysis of myxobacteria using 40 universal single-copy genes (gtp1, pheS, argS, rpsL, rpsG, rpsB, rplK, rplA, rplC, rplD, rplB, rplY, rpsC, rplN, rplE, rpsH, rplF, rpsE, rpsM, rpsK, rplM, rpsI, hisS, serS, rpsO, rpsS, rpsQ, rplP, rplO, cysS, rplR, leuS, rpsD, valS, tsaD, rpoB, rpoA, secY, ffh, and ftsY) which were identified as marker genes (MGs) using fetchMGs tool (http://motu-tool.org/fetchMG.html) [67]. Nucleotide sequences of these marker genes were retrieved from each genome, aligned using ClustalW, and further concatenated. The tree was generated using the GTR-GAMMA model of the ML method [bootstrap: 100] in RAxML (v8) tool and visualized by iTOL.
In silico DNA-DNA hybridization (DDH) and Average Nucleotide Identity (ANI) were calculated between M. rosea DSM 24000T and other 21 selected members (all representative genomes from suborder Sorangiineae and a few representative genomes from other families in order Myxoccales) using Genome-to-Genome Distance Calculator (GGDC) server [68] and ANI Calculator [69] respectively.
Working data, functional characterization, and estimation of orthologous genes
As two [Vulgatibacter incomptus, Pajaroellobacter abortibovis EBA] out of 21 selected genomes have relatively smaller genome size, 19 myxobacterial representatives from three suborders of the order Myxococcales, i.e. Sorangiineae (C. apiculatus DSM 436, P. fumosum, Polyangium sp. SDU3–1, S. cellulosum So ce26, S. cellulosum So ce56, S. cellulosum So ce836, S. cellulosum So ceGT47, S. cellulosum So0003–19-2, S. cellulosum So0007–03, S. cellulosum So0008–312, S. cellulosum So0157–2, S. cellulosum So0163, Labilithrix luteola DSM 27648T, S. amylolyticus DSM 53668T) [33], Cystobacterineae(A. gephyra DSM 2261T, C. fuscus DSM 52655, H. minutum DSM 14724T, Myxococcus hansupus), and Nannocystineae (E. salina DSM 1520) [9, 70,71,72] were selected to perform pangenome analysis via identifying homologous and orthologous proteins using Proteinortho (v6) (https://www.bioinf.uni-leipzig.de/Software/proteinortho/) [73]. Paralogous proteins in M. rosea were identified by all-against-all BLAST analysis (identity ≥30% and e-value ≤1e-10) of proteomes in M. rosea DSM 24000T using NCBI Blast+ 2.10.1 package [74]. Exogenous genetic materials in M. rosea DSM 24000T were identified by performing BLASTP (e-value ≤1e-30) against the dataset of plasmids, phages, and insertion sequence (IS) elements retrieved from the ACLAME database (http://aclame.ulb.ac.be/). Genomic islands in the M. rosea genome were identified using IslandViewer 4 [75].
Protein domains and functional analysis
Functional family and domains in all selected members of Sorangiineae were identified by scanning the Pfam-A database (v32.0) [76] using the hmmscan program (e-value ≤1e-5) of HMMER (http://hmmer.janelia.org/) [77]. Representative domains of two-component system (TCS) such as, HisKA (PF00512), Hpt (PF01627), HATPase_c (PF02518), His_kinase (PF06580), HWE_HK (PF07536), HisKA_2 (PF07568), HisKA_3 (PF07730), HATPase_c_2 (PF13581) and Response_reg (PF00072) were identified. Eukaryotic like kinases (Elks): Pkinase (PF00069), Pkinase_C (PF00433) and Pkinase_Tyr (PF07714); and protein phosphatases (PPs): PP2C_2 (PF13672, COG0631), SpoIIE (PF07228, COG2208), PPPs (PF00149, COG0639) DSPc (PF00782, COG2365) and LMWPc (PF01451, COG0394, COG2453), and PTPZ (COG4464) were explored. Functional categorization of M. rosea proteins was performed by estimating their Clusters of Orthologous Groups (COGs) [78] using the NCBI COG database [79]. The aforementioned gene clusters were grouped into various COG categories such as ‘Cellular processes and Signaling’ [CPS], ‘Information Storage and Processing’ [ISP], ‘Metabolism’ [MET], and ‘Poorly Characterized’ [PC] [80]. SignalP (v5.0) [81], PRED-TAT [http://www.compgen.org/tools/PREDTAT] and PRED-LIPO [http://www.compgen.org/tools/PRED-LIPO] were used to identify the secretome via signal peptide detection. Screened secretory protein sequences were used as queries on the TMHMM server, and protein sequences with 0–2 transmembrane domains were considered as final secretomes [82].
Estimation of biosynthetic gene clusters in M. rosea DSM 24000T
Prediction of BGCs in M. rosea was performed using the antiSMASH tool (v5.0) (https://antismash.secondarymetabolites.org) [83] and the identified BGCs were further processed using the BiG-SCAPE program (https://git.wageningenur.nl/medema-group/BiGSCAPE) [84]. Among the estimated BGCs, PUFA producing gene cluster was identified by considering the PUFA biosynthetic genes in Aetherobacter sp. SBSr008 (gene accession no. - AIJ50375.1, AIJ50376.1, and AIJ50377.1), A. fasciculatus SBSr002 (gene accession no. - AIJ50372.1, AIJ50373.1, and AIJ50374.1), and S. cellulosum So ce56 (gene accession no. - CAN90975.1, CAN90976.1, CAN90977.1, and CAN95221.1) [18]. BLAST searches were performed for each of the Pfa1, Pfa2, and Pfa3 protein sequences of M. rosea DSM 24000T, and were further considered for phylogenetic analysis using the WAG (G + I + F) model of the Maximum Likelihood method in MEGA X [85]. The trees were visualized using iTOL.
Availability of data and materials
The complete genome sequence of Minicystis rosea DSM 24000T and its annotations are deposited at DDBJ/ENA/GenBank under accession number CP016211.1.
Abbreviations
- CPS:
-
Cellular Processes and Signaling
- MET:
-
Metabolism
- ISP:
-
Information Storage and Processing
- TCS:
-
Two-component system
- HK:
-
Histidine kinases
- RR:
-
Response regulators
- ELKs:
-
Eukaryotic-like kinases
- PPs:
-
Protein phosphatases
- BGC:
-
Biosynthetic gene cluster
- PKS:
-
Polyketide synthase
- NRPS:
-
Nonribosomal polypeptide synthetase
- RiPP:
-
Ribosomal synthesized and post-translationally modified peptide
- PUFA:
-
Polyunsaturated fatty acid
- KS:
-
β-ketoacyl synthase
- MAT/AT:
-
Malonyl/acyltransferase
- ACP:
-
Acyl carrier protein
- KR:
-
Ketoreductase
- PS-DH:
-
PKS-like dehydratase
- CLF:
-
Chain length factor
- AT:
-
Acyltransferase
- DH:
-
Fab-A-like dehydratase
- DH’:
-
Pseudo-domain dehydratase
- AGPAT:
-
1-acylglycerol-3-phosphate O-acyltransferase
- PPTase:
-
4′-phosphopantetheinyl transferase
References
Dawid W. Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev. 2000;24(4):403–27. https://doi.org/10.1111/j.1574-6976.2000.tb00548.x.
Iizuka T, Jojima Y, Fudou R, Yamanaka S. Isolation of myxobacteria from the marine environment. FEMS Microbiol Lett. 1998;169(2):317–22. https://doi.org/10.1111/j.1574-6968.1998.tb13335.x.
Reichenbach H. The ecology of the myxobacteria. Environ Microbiol. 1999;1(1):15–21. https://doi.org/10.1046/j.1462-2920.1999.00016.x.
Sanford RA, Cole JR, Tiedje JM. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol. 2002;68(2):893–900. https://doi.org/10.1128/AEM.68.2.893-900.2002.
Wolf-Jackel GA, Hansen MS, Larsen G, Holm E, Agerholm JS, Jensen TK. Diagnostic studies of abortion in Danish cattle 2015-2017. Acta Vet Scand. 2020;62(1):1. https://doi.org/10.1186/s13028-019-0499-4.
Kaiser D, Robinson M, Kroos L. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb Perspect Biol. 2010;2(8):a000380. https://doi.org/10.1101/cshperspect.a000380.
Moine A, Agrebi R, Espinosa L, Kirby JR, Zusman DR, Mignot T, et al. Functional organization of a multimodular bacterial chemosensory apparatus. PLoS Genet. 2014;10(3):e1004164. https://doi.org/10.1371/journal.pgen.1004164.
Munoz-Dorado J, Marcos-Torres FJ, Garcia-Bravo E, Moraleda-Munoz A, Perez J. Myxobacteria: moving, killing, feeding, and surviving together. Front Microbiol. 2016;7:781.
Sharma G, Khatri I, Subramanian S. Comparative genomics of myxobacterial chemosensory systems. J Bacteriol. 2018;200(3):e00620.
Sharma G, Yao AI, Smaldone GT, Liang J, Long M, Facciotti MT, et al. Global gene expression analysis of the Myxococcus xanthus developmental time course. Genomics. 2021;113(1 Pt 1):120–34. https://doi.org/10.1016/j.ygeno.2020.11.030.
Thiery S, Kaimer C. The predation strategy of Myxococcus xanthus. Front Microbiol. 2020;11:2. https://doi.org/10.3389/fmicb.2020.00002.
Velicer GJ, Vos M. Sociobiology of the myxobacteria. Annu Rev Microbiol. 2009;63(1):599–623. https://doi.org/10.1146/annurev.micro.091208.073158.
Whitfield DL, Sharma G, Smaldone GT, Singer M. Peripheral rods: a specialized developmental cell type in Myxococcus xanthus. Genomics. 2020;112(2):1588–97. https://doi.org/10.1016/j.ygeno.2019.09.008.
Kiskowski MA, Jiang Y, Alber MS. Role of streams in myxobacteria aggregate formation. Phys Biol. 2004;1(3–4):173–83. https://doi.org/10.1088/1478-3967/1/3/005.
Gerth K, Pradella S, Perlova O, Beyer S, Muller R. Myxobacteria: proficient producers of novel natural products with various biological activities--past and future biotechnological aspects with the focus on the genus Sorangium. J Biotechnol. 2003;106(2–3):233–53. https://doi.org/10.1016/j.jbiotec.2003.07.015.
Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106(8):3468–96. https://doi.org/10.1021/cr0503097.
Reichenbach H, Hofle G. Biologically active secondary metabolites from myxobacteria. Biotechnol Adv. 1993;11(2):219–77. https://doi.org/10.1016/0734-9750(93)90042-L.
Gemperlein K, Rachid S, Garcia R, Wenzel S, Müller R. Polyunsaturated 1 fatty acid biosynthesis in myxobacteria: different PUFA synthases and their product diversity. Chem Sci. 2014;5(5):1733–41. https://doi.org/10.1039/c3sc53163e.
Lorente-Cebrian S, Costa AG, Navas-Carretero S, Zabala M, Martinez JA, Moreno-Aliaga MJ. Role of omega-3 fatty acids in obesity, metabolic syndrome, and cardiovascular diseases: a review of the evidence. J Physiol Biochem. 2013;69(3):633–51. https://doi.org/10.1007/s13105-013-0265-4.
Sahena F, Zaidul ISM, Jinap S, Saari N, Jahurul HA, Abbas KA, et al. PUFAs in fish: extraction, fractionation, importance in health. Compr Rev Food Sci F. 2009;8(2):59–74. https://doi.org/10.1111/j.1541-4337.2009.00069.x.
Shulse CN, Allen EE. Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One. 2011;6(5):e20146. https://doi.org/10.1371/journal.pone.0020146.
Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293(5528):290–3. https://doi.org/10.1126/science.1059593.
Allen EE, Bartlett DH. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology (Reading). 2002;148(Pt 6):1903–13. https://doi.org/10.1099/00221287-148-6-1903.
Morita N, Tanaka M, Okuyama H. Biosynthesis of fatty acids in the docosahexaenoic acid-producing bacterium Moritella marina strain MP-1. Biochem Soc Trans. 2000;28(6):943–5. https://doi.org/10.1042/bst0280943.
Orikasa Y, Nishida T, Hase A, Watanabe K, Morita N, Okuyama H. A phosphopantetheinyl transferase gene essential for biosynthesis of n-3 polyunsaturated fatty acids from Moritella marina strain MP-1. FEBS Lett. 2006;580(18):4423–9. https://doi.org/10.1016/j.febslet.2006.07.008.
Zhao JY, Zhong L, Shen MJ, Xia ZJ, Cheng QX, Sun X, et al. Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl Environ Microbiol. 2008;74(7):1980–7. https://doi.org/10.1128/AEM.02143-07.
Han K, Li ZF, Peng R, Zhu LP, Zhou T, Wang LG, et al. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci Rep. 2013;3(1):2101. https://doi.org/10.1038/srep02101.
Juhas M, Crook DW, Dimopoulou ID, Lunter G, Harding RM, Ferguson DJ, et al. Novel type IV secretion system involved in propagation of genomic islands. J Bacteriol. 2007;189(3):761–71. https://doi.org/10.1128/JB.01327-06.
van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19(9):479–84. https://doi.org/10.1016/S0168-9525(03)00203-8.
Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5(1):35. https://doi.org/10.1186/1471-2180-5-35.
Whitworth DE, Cock PJA. Two-component systems of the myxobacteria: structure, diversity and evolutionary relationships. Microbiology (Reading). 2008;154(Pt 2):360–72. https://doi.org/10.1099/mic.0.2007/013672-0.
Munoz-Dorado J, Inouye S, Inouye M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell. 1991;67(5):995–1006. https://doi.org/10.1016/0092-8674(91)90372-6.
Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25(11):1281–9. https://doi.org/10.1038/nbt1354.
Perez J, Castaneda-Garcia A, Jenke-Kodama H, Muller R, Munoz-Dorado J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci U S A. 2008;105(41):15950–5. https://doi.org/10.1073/pnas.0806851105.
Kole HK, Abdel-Ghany M, Racker E. Specific dephosphorylation of phosphoproteins by protein-serine and -tyrosine kinases. Proc Natl Acad Sci U S A. 1988;85(16):5849–53. https://doi.org/10.1073/pnas.85.16.5849.
Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75(1):192–212. https://doi.org/10.1128/MMBR.00042-10.
Madec E, Laszkiewicz A, Iwanicki A, Obuchowski M, Seror S. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol Microbiol. 2002;46(2):571–86. https://doi.org/10.1046/j.1365-2958.2002.03178.x.
Treuner-Lange A. The phosphatomes of the multicellular myxobacteria Myxococcus xanthus and Sorangium cellulosum in comparison with other prokaryotic genomes. PLoS One. 2010;5(6):e11164. https://doi.org/10.1371/journal.pone.0011164.
Goldman B, Bhat S, Shimkets LJ. Genome evolution and the emergence of fruiting body development in Myxococcus xanthus. PLoS One. 2007;2(12):e1329. https://doi.org/10.1371/journal.pone.0001329.
Steinegger M, Meier M, Mirdita M, Vohringer H, Haunsberger SJ, Soding J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics. 2019;20(1):473. https://doi.org/10.1186/s12859-019-3019-7.
Huntley S, Hamann N, Wegener-Feldbrugge S, Treuner-Lange A, Kube M, Reinhardt R, et al. Comparative genomic analysis of fruiting body formation in Myxococcales. Mol Biol Evol. 2011;28(2):1083–97. https://doi.org/10.1093/molbev/msq292.
Garcia R, Gemperlein K, Muller R. Minicystis rosea gen. nov., sp. nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium. Int J Syst Evol Microbiol. 2014;64(Pt 11):3733–42. https://doi.org/10.1099/ijs.0.068270-0.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. https://doi.org/10.1093/nar/28.1.27.
Konstantinidis K, Tiedje J. Trends between gene content and genome size in prokaryotic species with larger genomes. PNAS. 2004;101(9):3160–5. https://doi.org/10.1073/pnas.0308653100.
Bretscher AP, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133(2):763–8. https://doi.org/10.1128/jb.133.2.763-768.1978.
Bhat S, Boynton TO, Pham D, Shimkets LJ. Fatty acids from membrane lipids become incorporated into lipid bodies during Myxococcus xanthus differentiation. PLoS One. 2014;9(6):e99622. https://doi.org/10.1371/journal.pone.0099622.
Bode HB, Zeggel B, Silakowski B, Wenzel SC, Reichenbach H, Muller R. Steroid biosynthesis in prokaryotes: identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol Microbiol. 2003;47(2):471–81. https://doi.org/10.1046/j.1365-2958.2003.03309.x.
Hooper SD, Berg OG. On the nature of gene innovation: duplication patterns in microbial genomes. Mol Biol Evol. 2003;20(6):945–54. https://doi.org/10.1093/molbev/msg101.
Gevers D, Vandepoele K, Simillon C, Van de Peer Y. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 2004;12(4):148–54. https://doi.org/10.1016/j.tim.2004.02.007.
Hahn M. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100(5):605–17. https://doi.org/10.1093/jhered/esp047.
Cimermancic P, Medema M, Claesen J, Kurita K, Wieland Brown L, Mavrommatis K, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–21. https://doi.org/10.1016/j.cell.2014.06.034.
Heath RJ, Rock CO. The claisen condensation in biology. Nat Prod Rep. 2002;19(5):581–96. https://doi.org/10.1039/b110221b.
Tang Y, Tsai SC, Khosla C. Polyketide chain length control by chain length factor. J Am Chem Soc. 2003;125(42):12708–9. https://doi.org/10.1021/ja0378759.
Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18(4):380–416. https://doi.org/10.1039/a909079g.
Yoshida K, Hashimoto M, Hori R, Adachi T, Okuyama H, Orikasa Y, et al. Bacterial long-chain polyunsaturated fatty acids: their biosynthetic genes, functions, and practical use. Mar Drugs. 2016;14(5):94.
Ujihara T, Nagano M, Wada H, Mitsuhashi S. Identification of a novel type of polyunsaturated fatty acid synthase involved in arachidonic acid biosynthesis. FEBS Lett. 2014;588(21):4032–6. https://doi.org/10.1016/j.febslet.2014.09.023.
Garcia R, Stadler M, Gemperlein K, Muller R. Aetherobacter fasciculatus gen. nov., sp. nov. and Aetherobacter rufus sp. nov., novel myxobacteria with promising biotechnological applications. Int J Syst Evol Microbiol. 2016;66(2):928–38. https://doi.org/10.1099/ijsem.0.000813.
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–9. https://doi.org/10.1038/nmeth.2474.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. https://doi.org/10.1186/1471-2164-9-75.
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–8. https://doi.org/10.1093/nar/gkm160.
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. https://doi.org/10.1093/nar/25.5.955.
Gruber AR, Findeiss S, Washietl S, Hofacker IL, Stadler PF. RNAz 2.0: improved noncoding RNA detection. Pac Symp Biocomput. 2010;69–79. https://doi.org/10.1142/9789814295291_0011.
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):402. https://doi.org/10.1186/1471-2164-12-402.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. https://doi.org/10.1093/bioinformatics/btm404.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. https://doi.org/10.1093/bioinformatics/btu033.
Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39(Web Server issue):W475–8.
Milanese A, Mende DR, Paoli L, Salazar G, Ruscheweyh HJ, Cuenca M, et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat Commun. 2019;10(1):1014. https://doi.org/10.1038/s41467-019-08844-4.
Auch AF, von Jan M, Klenk HP, Goker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2(1):117–34. https://doi.org/10.4056/sigs.531120.
Yoon SH, Ha SM, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110(10):1281–6. https://doi.org/10.1007/s10482-017-0844-4.
Sharma G, Khatri I, Subramanian S. Complete genome of the starch-degrading myxobacteria Sandaracinus amylolyticus DSM 53668T. Genome Biol Evol. 2016;8(8):2520–9. https://doi.org/10.1093/gbe/evw151.
Sharma G, Narwani T, Subramanian S. Complete genome sequence and comparative genomics of a novel myxobacterium Myxococcus hansupus. PLoS One. 2016;11(2):e0148593. https://doi.org/10.1371/journal.pone.0148593.
Sharma G, Subramanian S. Unravelling the complete genome of Archangium gephyra DSM 2261T and evolutionary insights into myxobacterial chitinases. Genome Biol Evol. 2017;9(5):1304–11. https://doi.org/10.1093/gbe/evx066.
Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. https://doi.org/10.1093/nar/25.17.3389.
Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing G, Lau BY, Hoad G, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–5. https://doi.org/10.1093/nar/gkx343.
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. https://doi.org/10.1093/nar/gkt1223.
Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7(10):e1002195. https://doi.org/10.1371/journal.pcbi.1002195.
Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–6. https://doi.org/10.1093/nar/28.1.33.
Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43(Database issue):D261–9. https://doi.org/10.1093/nar/gku1223.
Hsiao WW, Ung K, Aeschliman D, Bryan J, Finlay BB, Brinkman FS. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 2005;1(5):e62. https://doi.org/10.1371/journal.pgen.0010062.
Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37(4):420–3. https://doi.org/10.1038/s41587-019-0036-z.
Mastronunzio JE, Tisa LS, Normand P, Benson DR. Comparative secretome analysis suggests low plant cell wall degrading capacity in Frankia symbionts. BMC Genomics. 2008;9(1):47. https://doi.org/10.1186/1471-2164-9-47.
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47(W1):W81–7. https://doi.org/10.1093/nar/gkz310.
Navarro-Munoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16(1):60–8. https://doi.org/10.1038/s41589-019-0400-9.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. https://doi.org/10.1093/molbev/msy096.
Acknowledgments
We thank Dr. Ramya TNC for providing us workplace to purify the genomic DNA and for proofreading the manuscript.
Funding
S.P. would like to thank the Science and Engineering Research Board, Department of Science and Technology for financial assistance under the National Post-Doctoral Fellowship (File Number- PDF/2019/003065). G.S. would like to thank the Council of Scientific and Industrial Research (CSIR) research fellowship and Department of Science and Technology (DST)-INSPIRE Faculty Award, Government of India for financial support. This work is supported by intramural funds of CSIR (OLP0175) and a project “Expansion and modernization of Microbial Type Culture Collection and Gene Bank (MTCC) jointly supported by the CSIR Grant No. BSC0402 and Department of Biotechnology (DBT) Govt. of India Grant No. BT/PR7368/INF/22/177/2012”. The funding bodies played no role in the design of the study, collection, analysis, and interpretation of data, and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
SP analyzed, investigated, validate, and prepared the manuscript; GS conceptualized, analyzed, investigated, supervised, reviewed, and edited the manuscript; SS: the project administrator, conceptualized, supervised, reviewed, and edited the manuscript. All authors have approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors with listed names declare no conflict of interest to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
Single-copy genes-based phylogenetic tree of myxobacteria. Branch color and leaf stripes represent the suborder and family-level taxonomy (color-coded), respectively.
Additional file 2: Fig. S2.
COG functional categorization of the Total, Accessory, Core, and Unique proteins in M. rosea. CPS = Cellular Processes and Signaling, ISP = Information Storage and Processing, MET = Metabolism, and PC = Poorly characterized.
Additional file 3: Table S1.
DDH and ANI values between M. rosea and selected myxobacteria. Color intensity changes from green to orange corresponding with higher to lower values, respectively.
Additional file 4: Table S2.
a) Distribution of core, unique, duplicate, ELK, BGC genes in M. rosea. Unique exogenous genes in Column E have been shaded using orange color.; b) Identified genomic islands in M. rosea genome; and c) Comparative distribution of Pfam families among the selected myxobacterial genomes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pal, S., Sharma, G. & Subramanian, S. Complete genome sequence and identification of polyunsaturated fatty acid biosynthesis genes of the myxobacterium Minicystis rosea DSM 24000T. BMC Genomics 22, 655 (2021). https://doi.org/10.1186/s12864-021-07955-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-07955-x