Abstract
“Burkholderia sprentiae” strain WSM5005T is an aerobic, motile, Gram-negative, non-spore-forming rod that was isolated in Australia from an effective N2-fixing root nodule of Lebeckia ambigua collected in Klawer, Western Cape of South Africa, in October 2007. Here we describe the features of “Burkholderia sprentiae” strain WSM5005T, together with the genome sequence and its annotation. The 7,761,063 bp high-quality-draft genome is arranged in 8 scaffolds of 236 contigs, contains 7,147 protein-coding genes and 76 RNA-only encoding genes, and is one of 20 rhizobial genomes sequenced as part of the DOE Joint Genome Institute 2010 Community Sequencing Program.
Similar content being viewed by others
Introduction
Legumes of the Fabaceae family of flowering plants have the unique capacity to form a symbiotic N2-fixing symbiosis with soil-inhabiting root nodule bacteria (RNB). This symbiosis supplies leguminous species with the essential bioavailable nitrogen that could otherwise not be obtained from soils that are inherently infertile. The agricultural region of south-west Western Australia contains such impoverished soils and the successful establishment of effective legume-RNB symbioses has been exploited to drive plant and animal productivity in this landscape without the reliance on nitrogenous fertilizer [1,2]. This landscape’s rainfall patterns appear to be changing, from a dry Mediterranean-type distribution to a generally reduced annual rainfall with a less predictable distribution [3]. Due to changes in rainfall patterns, the reproduction of the commercially used annual legume species is challenged. Perennial species might be more able to adapt to climate change, though few commercial perennial forage legumes are adapted to the acid and infertile soils encountered in the region [2]. Therefore, deep-rooted herbaceous perennial legumes including Rhynchosia and Lebeckia species adapted to acid and infertile soils have been investigated for use in this Australian agricultural setting [2,4,5]. The genus Lebeckia Thunb. is part of the Crotalarieae tribe, and refers to a group of 33 species of papilionoid legumes that are endemic to the southern and western parts of South Africa, which have similar soil and climate conditions to Western Australia [6,7]. This genus has recently been revised and is now subdivided into several sections, including Lebeckia s.s., Calobota and Wiborgiella [7]. The Lebeckia s.s. section, which includes L. ambigua, can easily be distinguished from other species by their acicular leaves and 5+5 anther arrangement [7–9].
In four expeditions to the Western Cape of South Africa, between 2002 and 2007, nodules and seeds of Lebeckia ambigua were collected and stored [5]. The isolation of RNB from these nodules gave rise to a collection of 23 microsymbionts that clustered into five groups within the genus Burkholderia [5]. Unlike most of the previously studied rhizobial Burkholderia strains, this South African group appears to be associated with papilionoid forage legumes (rather than Mimosa spp.). One of these Burkholderia strains has now been designated as the type strain of the new species “Burkholderia sprentiae” strain WSM5005T [10]. This isolate effectively nodulates Lebeckia ambigua and L. sepiaria [5]. Here we present a summary classification and a set of general features for “Burkholderia sprentiae” strain WSM5005T together with the description of the complete genome sequence and its annotation.
Classification and general features
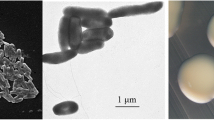
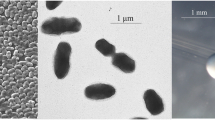
“Burkholderia sprentiae” strain WSM5005T is a motile, Gram-negative, non-spore-forming rod (Figure 1, left and center panels) in the order Burkholderiales of the class Betaproteobacteria [10]. It is fast growing, forming 2–4 mm diameter colonies within 2–3 days when grown on half Lupin Agar (½LA) [11] at 28°C. Colonies on ½LA are white-opaque, slightly domed, moderately mucoid with smooth margins (Figure 1, right panel).
Minimum Information about the Genome Sequence (MIGS) is provided in Table 1. Figure 2 shows the phylogenetic relationship of “Burkholderia sprentiae” strain WSM5005T in a 16S rRNA sequence based tree. This strain clusters closest to Burkholderia tuberum STM678T (CIP 108238T) and Burkholderia kururiensis KP23T with 98.2% and 96.9% sequence identity, respectively.
Phylogenetic tree showing the relationships of “Burkholderia sprentiae” strain WSM5005T (shown in blue print) with some of the bacteria in the order Burkholderiales based on aligned sequences of the 16S rRNA gene (1,322 bp internal region). All sites were informative and there were no gap-containing sites. Phylogenetic analyses were performed using MEGA, version 5.05 [25]. The tree was built using the maximum likelihood method with the General Time Reversible model. Bootstrap analysis [26] with 500 replicates was performed to assess the support of the clusters. Type strains are indicated with a superscript T. Strains with a genome sequencing project registered in GOLD [27] are in bold print and the GOLD ID is mentioned after the accession number. Published genomes are designated with an asterisk.
Symbiotaxonomy
“Burkholderia sprentiae” strain WSM5005T is part of a cadre of Burkholderia strains that were assessed for nodulation and nitrogen fixation on three separate L. ambigua genotypes (CRSLAM-37, CRSLAM-39 and CRSLAM-41) and on L. sepiaria [5]. Representatives of this group of nodule bacteria are generally Nod+ and Fix− on Macroptillium atropurpureum and appear to have a very narrow host range for symbiosis. They belong to a group of Burkholderia strains that nodulate papilionoid forage legumes rather than the classical Burkholderia hosts Mimosa spp. (Mimosoideae) [28].
Genome sequencing and annotation information
Genome project history
This organism was selected for sequencing on the basis of its environmental and agricultural relevance to issues in global carbon cycling, alternative energy production, and biogeochemical importance, and is part of the Community Sequencing Program at the U.S. Department of Energy, Joint Genome Institute (JGI) for projects of relevance to agency missions. The genome project is deposited in the Genomes OnLine Database [27] and an improved-high-quality-draft genome sequence in IMG. Sequencing, finishing and annotation were performed by the JGI. A summary of the project information is shown in Table 2.
Growth conditions and DNA isolation
“Burkholderia sprentiae” strain WSM5005T was grown to mid logarithmic phase in TY rich medium [29] on a gyratory shaker at 28°C. DNA was isolated from 60 mL of cells using a CTAB (Cetyl trimethyl ammonium bromide) bacterial genomic DNA isolation method [30].
Genome sequencing and assembly
The genome of “Burkholderia sprentiae” strain WSM5005T was sequenced at the Joint Genome Institute (JGI) using a combination of Illumina [31] and 454 technologies [32]. An Illumina GAii shotgun library which generated 76,247,610 reads totaling 5,794.8 Mb, and a paired end 454 library with an average insert size of 13 kb which generated 612,483 reads totaling 112.9 Mb of 454 data were generated for this genome. All general aspects of library construction and sequencing performed at the JGI can be found at [30]. The initial draft assembly contained 420 contigs in 8 scaffolds. The 454 paired end data was assembled with Newbler, version 2.3. The Newbler consensus sequences were computationally shredded into 2 kb overlapping fake reads (shreds). Illumina sequencing data were assembled with VELVET, version 1.0.13 [33], and the consensus sequences were computationally shredded into 1.5 kb overlapping fake reads (shreds). We integrated the 454 Newbler consensus shreds, the Illumina VELVET consensus shreds and the read pairs in the 454 paired end library using parallel phrap, version SPS - 4.24 (High Performance Software, LLC). The software Consed [34–36] was used in the following finishing process. Illumina data was used to correct potential base errors and increase consensus quality using the software Polisher developed at JGI (Alla Lapidus, unpublished). Possible mis-assemblies were corrected using gapResolution (Cliff Han, unpublished), Dupfinisher [37], or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR (J-F Cheng, unpublished) primer walks. A total of 352 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. The estimated genome size is 7.8 Mb and the final assembly is based on 65.2 Mb of 454 draft data which provides an average 8.4× coverage of the genome and 2,340 Mb of Illumina draft data which provides an average 300× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [38] as part of the DOE-JGI Annotation pipeline [39], followed by a round of manual curation using the JGI GenePRIMP pipeline [40]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. These data sources were combined to assert a product description for each predicted protein. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [41], RNAMMer [42], Rfam [43], TMHMM [44], and SignalP [45]. Additional gene prediction analyses and functional annotation were performed within the Integrated Microbial Genomes (IMG-ER) platform [46].
Genome properties
The genome is 7,761,063 nucleotides with 63.18% GC content (Table 3) and comprised of 8 scaffolds of 236 contigs. From a total of 7,223 genes, 7,147 were protein encoding and 76 RNA only encoding genes. Within the genome, 377 pseudogenes were also identified. The majority of genes (76.16%) were assigned a putative function whilst the remaining genes were annotated as hypothetical. The distribution of genes into COGs functional categories is presented in Table 4, Figure 3 and Figure 4.
Graphical map of the chromosome of “Burkholderia sprentiae” strain WSM5005T. From the bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Color code for Figure 3.
References
Herridge DF, Peoples MB, Boddey RM. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 2008; 311:1–18. http://dx.doi.org/10.1007/s11104-008-9668-3
Howieson JG, Yates RJ, Foster K, Real D, Besier B. Prospects for the future use of legumes. In: Dilworth MJ, James EK, Sprent JI, Newton WE, editors. Leguminous Nitrogen-Fixing Symbioses. London, UK: Elsevier; 2008. p 363–394.
George RJ, Speed RJ, Simons JA, Smith RH, Ferdowsian R, Raper GP, Bennett DL. Long-term groundwater trends and their impact on the future extent of dryland salinity in Western Australia in a varibale climate. Salinity Forum 2008. 2008.
Garau G, Yates RJ, Deiana P, Howieson JG. Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem 2009; 41:125–134. http://dx.doi.org/10.1016/j.soilbio.2008.10.011
Howieson JG, De Meyer SE, Vivas-Marfisi A, Ratnayake S, Ardley JK, Yates RJ. Novel Burkholderia bacteria isolated from Lebeckia ambigua — a perennial suffrutescent legume of the fynbos. Soil Biol Biochem 2013; 60:55–64. http://dx.doi.org/10.1016/j.soilbio.2013.01.009
Boatwright JS, Wink M, van Wyk BE. The generic concept of Lotononis (Crotalarieae, Fabaceae): Reinstatement of the genera Euchlora, Leobordea and Listia and the new genus Ezoloba. Taxon 2011; 60:161–177.
le Roux MM, Van Wyk BE. A revision of Lebeckia sect. Lebeckia: The L. pauciflora and L. wrightii groups (Fabaceae, Crotalarieae). S Afr J Bot 2009; 75:83–96. http://dx.doi.org/10.1016/j.sajb.2008.08.002
le Roux MM, Van Wyk BE. A revision of Lebeckia sect. Lebeckia: The L. sepiaria group. S Afr J Bot 2007; 73:118–130. http://dx.doi.org/10.1016/j.sajb.2006.09.005
Le Roux MM, Van Wyk BE. A revision of Lebeckia sect. Lebeckia: The L. plukenetiana group (Fabaceae, Crotalarieae). S Afr J Bot 2008; 74:660–676. http://dx.doi.org/10.1016/j.sajb.2008.04.005
De Meyer SE, Cnockaert M, Ardley J, Maker G, Yates RJ, Howieson JG, Vandamme P. Burkholderia sprentiae sp. nov. isolated from Lebeckia ambigua root nodules from South Africa. Int J Syst Evol Microbiol (In press).
Howieson JG, Ewing MA, D’antuono MF. Selection for acid tolerance in Rhizobium meliloti. Plant Soil 1988; 105:179–188. http://dx.doi.org/10.1007/BF02376781
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen M, Angiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed http://dx.doi.org/10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed http://dx.doi.org/10.1073/pnas.87.12.4576
Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1.
Validation List No. 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1–6. PubMed http://dx.doi.org/10.1099/ijs.0.64188-0
Garrity GM, Bell JA, Lilburn T. Class II. Betaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575.
Garrity GM, Bell JA, Lilburn T. Order I. Burkholderiales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575.
Garrity GM, Bell JA, Lilburn T. Family I. Burkholderiaceae fam. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575.
Editor L. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 45. Int J Syst Bacteriol 1993; 43:398–399. http://dx.doi.org/10.1099/00207713-43-2-398
Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol 1992; 36:1251–1275. PubMed http://dx.doi.org/10.11348-0421.1992.tb02129.x
Gillis M, Van TV, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 1995; 45:274–289. http://dx.doi.org/10.1099/00207713-45-2-274
Chen WX, Wang ET, Kuykendall LD. The Proteobacteria. New York: Springer-Verlag; 2005.
Agents B. Technical rules for biological agents. TRBA (http://www.baua.de):466.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. PubMed http://dx.doi.org/10.1038/75556
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011; 28:2731–2739. PubMed http://dx.doi.org/10.1093/molbev/msr121
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39:783–791. http://dx.doi.org/10.2307/2408678
Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475–D479. PubMed http://dx.doi.org/10.1093/nar/gkm884
Elliott GN, Chou JH, Chen WM, Bloemberg GV, Bontemps C, Martínez-Romero E, Velázquez E, Young JPW, Sprent JI, James EK. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ Microbiol 2009; 11:762–778. PubMed http://dx.doi.org/10.1111/j.1462-2920.2008.01799.x
Reeve WG, Tiwari RP, Worsley PS, Dilworth MJ, Glenn AR, Howieson JG. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology 1999; 145:1307–1316. PubMed http://dx.doi.org/10.1099/13500872-145-6-1307
DOE Joint Genome Institute. http://my.jgi.doe.gov/general/index.html
Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433–438. PubMed http://dx.doi.org/10.1517/14622416.5.4.433
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:376–380. PubMed
Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Current Protocols in Bioinformatics 2010; Chapter 11:Unit 11 5.
Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998; 8:175–185. PubMed http://dx.doi.org/10.1101/gr.8.3.175
Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175–185. PubMed http://dx.doi.org/10.1101/gr.8.3.175
Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195–202. PubMed http://dx.doi.org/10.1101/gr.8.3.195
Han C, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Valafar HRAH, editor. Proceeding of the 2006 international conference on bioinformatics & computational biology: CSREA Press; 2006. p 141–146.
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119. PubMed http://dx.doi.org/10.1186/1471-2105-11-119
Mavromatis K, Ivanova NN, Chen IM, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci 2009; 1:63–67. PubMed http://dx.doi.org/10.4056/sigs.632
Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455–457. PubMed http://dx.doi.org/10.1038/nmeth.1457
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955–964. PubMed
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100–3108. PubMed http://dx.doi.org/10.1093/nar/gkm160
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res 2003; 31:439–441. PubMed http://dx.doi.org/10.1093/nar/gkg006
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567–580. PubMed http://dx.doi.org/10.1006/jmbi.2000.4315
Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783–795. PubMed http://dx.doi.org/10.1016/j.jmb.2004.05.028
Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271–2278. PubMed http://dx.doi.org/10.1093/bioinformatics/btp393
Acknowledgements
This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. We gratefully acknowledge the funding received from the Murdoch University Strategic Research Fund through the Crop and Plant Research Institute (CaPRI) and the Centre for Rhizobium Studies (CRS) at Murdoch University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Reeve, W., De Meyer, S., Terpolilli, J. et al. Genome sequence of the Lebeckia ambigua-nodulating “Burkholderia sprentiae” strain WSM5005T. Stand in Genomic Sci 9, 385–394 (2013). https://doi.org/10.4056/sigs.4558268
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.4558268