Abstract
The genus Amphibacillus was established in 1990, and seven additional species were described in the past two decades. Amphibacillus jilinensis Y1T is a facultatively anaerobic and alkaliphilic bacterium isolated from a soda lake in China. Here we describe the structural and genetic features of the draft genome about the type strain Y1T (3,831,075 bp, with a G+C content of 37.27%). This is the first genome report of the Amphibacillus genus.
Similar content being viewed by others
Introduction
The genus Amphibacillus belongs to the family Bacillaceae and was established in 1990 [1]. Currently the genus comprises eight validly published species: A. xylanus [1], A. indicireducens [2], A. cookii [3], A. marinus [4], A. jilinensis [5], A. sediminis [6], A. fermentum and A. tropicus [7]. All are Gram-positive, moderately alkaliphilic, facultatively anaerobic rods [5,6]. All can grow at pH 9.0 and one can grow at pH 12.0 [2–4,6]. Amphibacillus jilinensis Y1T (=CGMCC 1.5123T =JCM 16149T) was isolated from a soda lake in Jilin province, China, and grows at pH range from 7.5 to 10.5 with an optimum at 9.0 [5]. Strain Y1T can utilize a large spectrum of substrates as sources of carbon and energy, can grow both aerobically and anaerobically, and tolerate Na+ up to 2.8 M. In this genus, three species have been sequenced. A finished genome sequence is Amphibacillus xylanus NBRC 15112 (NCBI Accession Number AP012050) and two incomplete sequences are A. jilinensis Y1T (NCBI Accession Number AMWI00000000) and Amphibacillus sediminis Shu-P-Ggiii25-2 (NCBI BioProject ID PRJDB405) according to the GOLD records [8,9]. Here we report this draft genome of A. jilinensis Y1T, the first genome from genus Amphibacillus to be sequenced.
Classification and features
A sediment sample was collected from a soda lake (44°45′N, 123°34′E) in Jilin province, China, in November 2007. There is no freshwater river to flow into the lake. Atmospheric water and groundwater are the only water sources of this lake. The lake is rich in Na+ (257.2 mg/l), CO32- (50.7 mg/l), Cl- (10.1 mg/l), HCO3- (6.5 mg/l) and SO42- (4.4 mg/l), with the pH of the water sample in the same geographical location being 10.0 [5]. The strain Y1T was isolated from enrichment cultures of sediment sample by the Hungate roll-tube technique [10] under a gas phase of O2-free N2 [1,5].
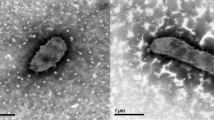
Comparative 16S rRNA gene sequence analysis by BLASTN [11,12] using the NCBI-NR/NT database revealed 93.4-98.8% sequence similarity to members of the genus Amphibacillus. Neighbor-Joining phylogenetic analysis based on Tamura-Nei model indicated the taxonomic status of strain Y1T is clearly classified into the same branch with genus Amphibacillus, and the most closely related genus is Halolactibacillus (Figure 1). A. jilinensis Y1T can tolerant high salinity but can also survive without Na+. Growth occurs under either aerobic or anaerobic conditions. The optimal growth condition of strain Y1T occurs in medium JY with 0.5 M Na+ (0.06 M NaHCO3 and 0.44 M NaCl) [5]. The optimum pH is 9.0, with a growth range of pH 7.5–10.5. No growth was observed at pH 7.0 or 11.0. Strain Y1T is mesophilic, with a temperature range of 15–45 ̱C and optimum growth at 32 ̱C [Table 1]. Cell morphology, motility and sporulation were examined by using transmission electron (H-600, Hitachi) microscopy. Cells of strain Y1T are straight rods with petritrichous flagella, which have a diameter ranging 0.4–0.6 µm and a length of 2.0–3.2 µm (Figure 2a). In the late-exponential and stationary phases of growth, the rods can form terminal endospores (Figure 2b).
Phylogenetic tree highlighting the position of A. jilinensis strain Y1T relative to other type strains within the Amphibacillus genus and with the relative Halolactibacillus genus. The strains and their corresponding Genbank accession numbers are shown following the organism name and indicated in parentheses. Three strains have their corresponding NCBI genome project IDs and sequencing status [8,13] listed here: PRJNA42371 of A. xylanus DSM 6626T, complete; PRJNA171498 of A. jilinensis Y1T, Draft; PRJDB405 of A. sediminis Shu-P-Ggiii25-2T, in progress. The phylogenetic tree uses 16S rRNA gene sequences aligned by the CLUSTALW [14], and phylogenetic inferences were made using Neighbor-joining method based on Tamura-Nei model within the MEGA5 software [15]. Numbers at the branching nodes are percentages of bootstrap values based on 1,000 replications. The scale bar indicates a 1% substitution per nucleotide position. Bacillus subtilis DSM 10T was used as an outgroup.
Genome sequencing information
Genome project history
The genome of A. jilinensis was selected for next-generation sequencing on the consideration of its facultatively anaerobic characterization and as a new member in genus Amphibacillus. This is the first genome report for any of the eight Amphibacillus species. Two others are the subject of ongoing own genome projects. This Whole Genome Shotgun project of A. jilinensis was deposited at DDBJ/EMBL/GenBank under the accession AMWI00000000 and consists of 83 contigs (further assembling constructed these contigs into 30 scaffolds). Table 2 presents the project information and its association with MIGS version 2.0 compliance [16].
Growth conditions and DNA isolation
A. jilinensis Y1T was cultivated aerobically in modified JY medium, which contains (per liter distilled water) 2.0 g yeast extract (Difco), 5.0 g sucrose, 0.2 g KCl, 0.2 g KH2PO4, 0.1 g MgCl2. 6H2O, 0.5 g NH4Cl, 0.1 g CaCl2, 0.06 M NaHCO3 and 0.44 M NaCl, final pH 9.0 at 32°C for 3 days [5]. Genomic DNA was extracted using the method described by Marmur [28]. The yield, purity and the concentration of genomic DNA was judged by the 0.7% agarose gel electrophoresis with λ-Hind III digest DNA Marker (TaKaRa, Dalian, China) and measured by the NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific Inc., USA). About 736.6 εg genomic DNA at the concentration 744 ng/εl was obtained.
Genome sequencing and assembly
Genomic DNA sequencing of A. jilinensis Y1T was performed using Solexa paired-end sequencing technology (HiSeq2000 system, Illumina, Inc., USA) [29] with a whole-genome shotgun (WGS) strategy, with a 500 bp-span paired-end library (∼500 Mb available reads, ∼130-fold genome coverage) and a 2,000 bp-span paired-end library (∼250 Mb available reads, ∼65-fold genome coverage). All these clean reads were assembled into 83 contigs (the minimum length is 231 bp) and 30 scaffolds (the minimum length is 542 bp) using the SOAPdenovo v.1.05 [30,31,50]. The quality of the sequencing reads data was estimated by G+C content and sequencing depth correlation analysis.
Genome annotation
The tRNAs and rRNAs were identified using tRNAscan-SE [32], RNAmmer [33] and Rfam database [34]; The open reading frames (ORFs) and the functional annotation of translated ORFs were predicted and achieved by using the RAST server online [35,51]. Classification of some predicted genes and pathways were analyzed using COGs [36,37] and KEGG [38–40] databases. Meanwhile, we used the InterPro [41,42] to obtain the GO annotation with the database of Pfam [43].
Genome properties
The draft genome sequence of A. jilinensis Y1T revealed a genome size of 3,836,603 bp (scaffold length) and a G+C content of 37.27%. These scaffolds contain 3,649 coding sequences (CDSs), 51 tRNAs (removed 3 Pseudo tRNAs) and incomplete rRNA operons (two 5 S rRNA and one 16 S rRNA). A total of 2,683 protein-coding genes (67.72%) were assigned a predicted function (Table 3) and genes have been categorized into COGs functional groups (Table 4).
Insights from the genome sequence
The genomic annotation results suggest that strain Y1T can adapt to an extremely basic environments. A large number of genes related to carbohydrate metabolism can encode proteins that provide a stable energy supply to maintain the lower internal pH despite the high external pH [44]. Several cation/proton antiporters were found in the genome, which are also crucial for the maintenance of internal pH [45]. However, the lower number of these genes in Y1T when compared to Bacillus pseudofirmus OF4 [44] may imply another way of importing protons into the cell. Meanwhile, as a facultatively anaerobic bacterium, 27 oxidative stress related genes are found in the predicted annotations, such as manganese superoxide dismutase (EC 1.15.1.1), superoxide dismutase [Cu-Zn] precursor (EC 1.15.1.1), organic hydroperoxide resistance transcriptional regulator and CoA-disulfide reductase (EC 1.8.1.14). For facultatively anaerobic strains, these superoxide dismutases (SODs) may be critical because the systems can help to regulate intracellular oxidative stress when the cells grow during aerobic respiration, and can also be used in the treatment of disease, study of pharmacological activity [46] and in the cosmetic industry. It also contains 34 two-component system genes that encode response regulators and sensor histidine kinases. The two-component systems appear to be used to respond to a wide variety of stimuli, including the presence of nutrients, antibiotics and chemoattractants in the environment, changes in osmolarity, temperature, pH, etc [47,48]. This is especially true in strain Y1T, in which these systems are thought to be used for recognizing environmental pH, and regulating its internal osmotic stress to survive various environments [49]. According to the database Pfam [43], there are also 9 CRISPRs-associated (Cas) proteins or Cas protein families in this genome of A. jilinensis.
Conclusion
Strain Y1T is the fifth member of the genus Amphibacillus to be described and is the first for which a genome sequence report is available. These data will provide a new perspective of how microorganisms adapt to anoxic and alkaline environments, and may also provide a pool of functional enzymes that work at higher pH.
References
Niimura Y, Koh E, Yanagida F, Suzuki KI, Komagata K, Kozaki M. Amphibacillus xylanus gen. nov., sp. nov., a facultatively anaerobic sporeforming xylan-digesting bacterium which lacks cytochrome, quinone, and catalase. Int J Syst Bacteriol 1990; 40:297–301. http://dx.doi.org/10.1099/00207713-40-3-297
Hirota K, Aino K, Nodasaka Y, Morita N, Yumoto I. Amphibacillus indicireducens sp. nov., a facultatively alkaliphile that reduces an indigo dye. Int J Syst Evol Microbiol 2013; 63:464–469. PubMed http://dx.doi.org/10.1099/ijs.0.037622-0
Pugin B, Blamey JM, Baxter BK, Wiegel J. Amphibacillus cookii sp. nov., a facultatively aerobic, sporeforming, moderate halophilic, alkalithermotolerant bacterium from Great Salt Lake, Utah. Int J Syst Evol Microbiol 2012; 62:2090–2096. PubMed http://dx.doi.org/10.1099/ijs.0.034629-0
Ren B, Yang N, Wang J, Ma XL, Wang Q, Xie F, Guo H, Liu ZH, Pugin BÆ, Zhang LX. Amphibacillus marinus sp. nov., a new member of the genus Amphibacillus isolated from the South China Sea. Int J Syst Evol Microbiol 2013; 63:1485–1491. PubMed http://dx.doi.org/10.1099/ijs.0.045807-0
Wu XY, Zheng G, Zhang WW, Xu XW, Wu M, Zhu XF. Amphibacillus jilinensis sp. nov., a facultatively anaerobic, alkaliphilic bacillus from a soda lake. Int J Syst Evol Microbiol 2010; 60:2540–2543. PubMed http://dx.doi.org/10.1099/ijs.0.018259-0
An SY, Ishikawa S, Kasai H, Goto K, Yokota A. Amphibacillus sediminis sp. nov., an endospore-forming bacterium isolated from lake sediment in Japan. Int J Syst Evol Microbiol 2007; 57:2489–2492. PubMed http://dx.doi.org/10.1099/ijs.0.64926-0
Zhilina T, Garnova E, Tourova T, Kostrikina N, Zavarzin G. Amphibacillus fermentum sp. nov. and Amphibacillus tropicus sp. nov., New Alkaliphilic, Facultatively Anaerobic, Saccharolytic Bacilli from Lake Magadi. Microbiology 2001; 70:825–837. PubMed
Pagani I, Liolios K, Jansson J, Chen IMA, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2012; 40(D1):D571–D579. PubMed http://dx.doi.org/10.1093/nar/gkr1100
Genomes On Line Database. http://www.genomesonline.org
Humane R. A roll tube method for cultivation of strict anaerobes. METHODS IN MICROBIOLOGY, VOLUME 3B 1969;3:117.
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res 2008; 36(suppl 2):W5. PubMed http://dx.doi.org/10.1093/nar/gkn201
McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 2004; 32(suppl 2):W20–W25. PubMed http://dx.doi.org/10.1093/nar/gkh435
Liolios K, Chen IMA, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38(suppl 1):D346–D354. PubMed http://dx.doi.org/10.1093/nar/gkp848
Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2002. PubMed http://dx.doi.org/10.1002/0471250953.bi0203s00
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011; 28:2731–2739. PubMed http://dx.doi.org/10.1093/molbev/msr121
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed http://dx.doi.org/10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed http://dx.doi.org/10.1073/pnas.87.12.4576
Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28:1–6. http://dx.doi.org/10.1099/00207713-28-1-1
Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119–169.
Murray RGE. The Higher Taxa, or, a Place for Everything…? In: Holt JG (ed), Bergey’s Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31–34.
List Editor. List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol 2010; 60:469–472. http://dx.doi.org/10.1099/ijs.0.022855-0
Ludwig W, Schleifer KH, Whitman WB. Class I. Bacilli class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 19–20.
Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225–420. http://dx.doi.org/10.1099/00207713-30-1-225
Prévot AR. In: Hauderoy P, Ehringer G, Guillot G, Magrou. J., Prévot AR, Rosset D, Urbain A (eds), Dictionnaire des Bactéries Pathogènes, Second Edition, Masson et Cie, Paris, 1953, p. 1–692.
Fischer A. Untersuchungen über bakterien. Jahrbücher für Wissenschaftliche Botanik 1895; 27:1–163.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. PubMed http://dx.doi.org/10.1038/75556
Gene Ontology project. http://www.geneontology.org/GO.evidence.shtml
Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 1962; 5:109–118. PubMed http://dx.doi.org/10.1016/S0022-2836(62)80066-7
Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008; 456:53–59. PubMed http://dx.doi.org/10.1038/nature07517
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 2010; 20:265–272. PubMed http://dx.doi.org/10.1101/gr.097261.109
Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics 2008; 24:713. PubMed http://dx.doi.org/10.1093/bioinformatics/btn025
Lowe TM, Eddy S. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955–964. PubMed
Lagesen K, Hallin P. Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100. PubMed http://dx.doi.org/10.1093/nar/gkm160
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res 2003; 31:439. PubMed http://dx.doi.org/10.1093/nar/gkg006
Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9:75. PubMed http://dx.doi.org/10.1186/1471-2164-9-75
Tatusov RL, Natale D, Garkavtsev I, Tatusova T, Shankavaram U, Rao B, Kiryutin B, Galperin M, Fedorova N, Koonin E. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 2001; 29:22–28. PubMed http://dx.doi.org/10.1093/nar/29.1.22
Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 2000; 28:33–36. PubMed http://dx.doi.org/10.1093/nar/28.1.33
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res 2008(36 Database):D480 — 484.
Moriya Y, Itoh M, Okuda S, Kanehisa M. KAAS: KEGG automatic annotation server. Genome Informatics 2005; 5:2005.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27. PubMed http://dx.doi.org/10.1093/nar/28.1.27
Zdobnov EM, Apweiler R. InterProScan Can integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001; 17:847. PubMed http://dx.doi.org/10.1093/bioinformatics/17.9.847
Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MDR. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 2001; 29:37. PubMed http://dx.doi.org/10.1093/nar/29.1.37
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res 2004; 32(suppl 1):138D. PubMed http://dx.doi.org/10.1093/nar/gkh121
Janto B, Ahmed A, Ito M, Liu J, Hicks DB, Pagni S, Fackelmayer OJ, Smith TA, Earl J, Elbourne LD, et al. Genome of alkaliphilic Bacillus pseudofirmus OF4 reveals adaptations that support the ability to grow in an external pH range from 7.5 to 11.4. Environ Microbiol 2011; 13:3289–3309. PubMed http://dx.doi.org/10.1111/j.1462-2920.2011.02591.x
Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. [table of contents.]. Microbiol Mol Biol Rev 1999; 63:735–750. PubMed
Muscoli C, Cuzzocrea S, Riley DP, Zweier JL, Thiemermann C, Wang ZQ, Salvemini D. On the selectivity of superoxide dismutase mimetics and its importance in pharmacological studies. Br J Pharmacol 2003; 140:445–460. PubMed http://dx.doi.org/10.1038/sj.bjp.0705430
Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 2002; 3:S3013. PubMed http://dx.doi.org/10.1186/gb-2002-3-10-reviews3013
Attwood PV, Piggott M, Zu X, Besant P. Focus on phosphohistidine. Amino Acids 2007; 32:145–156. PubMed http://dx.doi.org/10.1007/s00726-006-0443-6
Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni ME, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 2010; 64:539–559. PubMed http://dx.doi.org/10.1146/annurev.micro.112408. 134054
SOAP. denovo v.1.05. http://soap.genomics.org.cn/soapdenovo.html
RAST server online. http://rast.nmpdr.org
Acknowledgements
We thank Xiao-Yue Wu for her work in isolation and characterization of this new bacterial species and Xin-Qi Zhang for her professional advice. This work was supported by the Chinese Natural Science Foundation (grant no. 31170001) and Zhoushan Science and Technology Projects (no. 2012C33024 & no. 2011C31013).
Author information
Authors and Affiliations
Corresponding author
Additional information
The first two authors contributed equally to this work.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Cheng, H., Fang, MX., Jiang, XW. et al. Draft genome sequence of Amphibacillus jilinensis Y1T, a facultatively anaerobic, alkaliphilic and halotolerant bacterium. Stand in Genomic Sci 8, 491–499 (2013). https://doi.org/10.4056/sigs.4107829
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.4107829