Summary
Histidine protein kinases (HPKs) are a large family of signal-transduction enzymes that autophosphorylate on a conserved histidine residue. HPKs form two-component signaling systems together with their downstream target proteins, the response regulators, which have a conserved aspartate in a so-called 'receiver domain' that is phosphorylated by the HPK. Two-component signal transduction is prevalent in bacteria and is also widely used by eukaryotes outside the animal kingdom. The typical HPK is a transmembrane receptor with an amino-terminal extracellular sensing domain and a carboxy-terminal cytosolic signaling domain; most, if not all, HPKs function as dimers. They show little similarity to protein kinases that phosphorylate serine, threonine or tyrosine residues, but may share a distant evolutionary relationship with these enzymes. In excess of a thousand known genes encode HPKs, which are important for multiple functions in bacteria, including chemotaxis and quorum sensing, and in eukaryotes, including hormone-dependent developmental processes. The proteins divide into at least 11 subfamilies, only one of which is present in eukaryotes, suggesting that lateral gene transfer gave rise to two-component signaling in these organisms.
Similar content being viewed by others
Gene organization and evolutionary history
Histidine protein kinases (HPKs), together with their partner response regulators, are undoubtedly the most widely used of all signal-transduction enzymes in nature. They are present in all three major kingdoms of life (the Bacteria, Archaea and Eukarya) [1,2], and function in the sensing of a cell's external environment [3]. For unicellular organisms this usually equates to sensing nutrients, chemoattractants, osmotic conditions and so on. HPKs are also responsible for coordinating the behavior of cell populations. In bacteria this takes the form of quorum sensing [4]. In Eukarya, where HPKs appear to be confined to plants and to free-living organisms, such as yeasts, fungi and protozoa, HPKs have roles in regulating hormone-dependent developmental processes [5]; for example, HPKs are responsible for transducing the effects of the hormone ethylene in plants [6], and for coordinating the development of the fruiting body of the slime mold Dictyostelium discoideum [5]. No HPK (or response regulator) genes are present in the completed genome sequences of Caenorhabditis elegans [7], Drosophila melanogaster [8], or Homo sapiens [9], and it is thought that these enzymes are absent from the animal kingdom as a whole.
HPKs (and response regulators) probably arose in bacteria and, with few exceptions, are found in all bacterial species [2], which have a wide variation of HPK gene numbers: for example, the Escherichia coli and Bacillus subtilis genomes both contain about 25 HPK genes; in contrast, Helicobacter pylori has only four HPKs, and the three Mycoplasma species whose genomes have been sequenced (M. genitalium, M. pneumoniae, and M. pulmonis) are among the rare exceptions with no HPK genes. There are well over a thousand HPK genes currently in sequence databases, and HPK genes are found much less frequently in archaea and eukaryotes than in bacteria. HPK proteins can be divided by sequence analysis into 11 subfamilies, and virtually all eukaryotic HPKs belong to a single subfamily [1]. This, and the relative scarcity of HPK genes in archaea and eukaryotes, supports the notion that HPK (and response regulator) genes arrived in eukaryotes and archaea by lateral gene transfer from bacteria.
In prokaryotes, the genes for cognate pairs of HPKs and response regulators are typically found together in a single operon, such as the EnvZ-OmpR system that controls osmosensing in E. coli [3]. The functional relationship between these signaling partners is therefore reflected in their gene organization. Cross-regulation of a response regulator by additional histidine kinases does occur, however. For instance, the OmpR response regulator receives phosphate in vivo not only from its cognate HPK EnvZ, but also from the ArcB histidine kinase [10], although the ArcB and OmpR genes are not on the same operon.
In eukaryotes, HPK and receiver domains, containing the aspartate that is phosphorylated by the HPK, are generally encoded within a single gene, whereas in bacteria the receiver domain is part of the response regulator. Eukaryotic HPK proteins therefore contain both elements of the traditional two-component pathway, and are referred to as 'hybrid kinases'. Such hybrid kinases, while predominating in eukaryotes, are only a small minority among prokaryotic HPKs; all hybrid HPKs (both eukaryotic and prokaryotic) belong to a subdivision of the HPK1 gene family, one of the 11 HPK families (Figure 1) [1].
Conserved sequence motifs in the 11 histidine protein kinase subfamilies. (a) Motifs representative of conserved sequences in the HPK1 family, which includes a majority of all HPKs, including all eukaryotic HPKs [1]. (b) The relative positions of the conserved motifs in a representative HPK (E. coli EnvZ). Regions known or predicted to be α helical are shown as rectangles, and β sheets as arrows [14,21]. The conserved regions that make up the HPK core, designated the H, N, D, F and G boxes (shown in blue), typically span approximately 200 residues in total [1,19]. HAMP is a linker domain, and TM1 and TM2 are transmembrane helices. Sequence alignments of (c) the H box, (d) the N box, (e) the D and F boxes, and (f) the G box for one representative member of each of the 11 HPK subfamilies (except for the HPK1 family, where one member of HPK1a and one member of HPK1b are shown). The proteins used to make this alignment were: HPK1a, Pseudomonas aeruginosa KinB (595 amino acids in total); HPK1b, E. coli TorS (914 amino acids); HPK2, E. coli EnvZ (450 amino acids); HPK3, E. coli PhoQ (486 amino acids); HPK4, E. coli NtrB (349 amino acids); HPK5, E. coli DcuS (543 amino acids); HPK6, Archaeoglobus fulgidus g2648416 (607 amino acids); HPK7, E. coli NarQ (566 amino acids); HPK8, E. coli YehU (561 amino acids); HPK9, E. coli CheA (654 amino acids); HPK10, Streptococcus pneumoniae ComD (441 amino acids); HPK11, Methanobacterium mth292 (564 amino acids). The regions chosen for these alignments were taken from subfamily-specific alignments carried out in one of our previous studies [1], and are indicated in parentheses. Sequences were aligned using ClustalW. To simplify the alignment, a 25 amino-acid region of HPK9 was removed in the D-box region (indicated by an open square), and a 3 amino-acid region of HPK8 was removed in the G-box region (indicated by a filled square). HPK9 does not contain an H box in its dimerization domain, so HPK9 was left out of the H-box alignment. HPK7 and HPK8 do not contain an F box. The HPK11 subfamily contains a partially conserved F box, but the example used for this alignment does not. Highly conserved residues (those present in a majority of the subfamilies) are shown boxed in dark blue, and a consensus sequence is presented below the aligned sequences; positions containing chemically similar residues are shown boxed in light blue and indicated by a dot in the consensus sequence.
Characteristic structural features
HPKs catalyze the transfer of phosphate from ATP to a unique histidine residue, and all HPKs have a conserved ATP-binding catalytic domain that is required for kinase activity. This catalytic domain, together with a dimerization domain, forms the kinase core. The HPKs are classified into 11 subfamilies on the basis of the sequences of these two core domains [1]. Figure 1 shows a sequence alignment of the core domains with a representative of each subfamily.
The core domains
Histidine kinase activity depends on homodimer formation, with the dimerization domains, which have two-stranded coiled-coils, coming together to form a four-helix bundle [11,12]. It can be seen in Figure 1 that, except for the hpk9 family (the CheA family), the dimerization domain includes a motif, known as the H-box, which contains the site of autophosphorylation. As in the case of tyrosine protein kinases, such as the insulin receptor, HPK-mediated autophosphorylation appears to occur in trans, with the catalytic domain of one subunit in a dimer phosphorylating the H-box histidine in the opposing subunit [3]. Many HPKs also have phosphatase activity which dephosphorylates the response regulator and opposes kinase function [3, 13] (for details see the Mechanism section); phosphatase activity is mediated by the dimerization domain in these HPKs.
The catalytic domain of HPKs has clear sequence and structural homology to the ATP-binding domains of type II topoisomerases (such as GyrB), the mismatch repair protein MutL, and the heat-shock protein Hsp90 [14,15,16]. These domains form a family with a conserved structure of several α helices packed over one face of a large, mostly antiparallel, β sheet, forming a loop that closes over the bound ATP (the 'ATP lid'). The ATP lid and the entire ATP-binding site are poorly organized in the absence of ATP or ATP analogs [14,15,17]. A substantial ordering of structure and other conformational changes in the ATP lid can be seen in the X-ray crystal structures of nucleotide-bound forms of MutL and GyrB as well as the HPKs CheA (which is involved in bacterial chemotaxis) [16] and PhoQ (which senses environmental concentrations of Ca2+ and Mg2+) [18]. Within the HPK catalytic domain four conserved motifs, the N, D, F, and G boxes, are involved in ATP binding [16,18], and probably also in catalysis and phosphotransfer. The motifs can be seen in Figure 1. Some subfamilies (HPK7 and HPK8) appear to have no identifiable F box, and the HPK10 subfamily has no identifiable D box. The D and G boxes [1,19] have also been labeled the G1 and G2 boxes, respectively [20].
Figure 2 shows representations of the only three available structures of HPKs, from which a number of insights have been gained about the overall structure of HPKs [14,15,21], as well as the role of conserved residues of the catalytic domain in interactions with the Mg2+ co-factor, which is required for ATP binding, and the nucleotide substrate [14,16,18]. The aspartate that defines the D box is hydrogen-bonded directly to the adenine ring of the ATP. Buried water molecules form hydrogen bonds that connect additional amino-acid residues in the N and D boxes to the adenine. The bound Mg2+ bridges from the nucleotide phosphates to residues in the N box. The F box is part of the ATP lid, whereas the G box forms the flexible hinge at the end of the ATP lid. Hydrolysis of ATP is coupled to Mg2+ release and conformational changes in the ATP-binding cavity, and the ATP lid does not remain wellordered with ADP in the binding site. The ordering of the ATP lid may couple ATP binding to interactions between domains, such as those between the catalytic domain and the H-box region. On the basis of the CheA structure, it seems that there is substantial flexibility in the region of the hinge that joins the dimerization and catalytic domains [15]. This flexibility maybe critical for the domain rearrangements that occur during the catalytic cycle [22].
A comparison of the three-dimensional structures of three HPK families (HPK2, HPK3, and HPK9) shows the strong conservation of the catalytic domain structure. (a) The kinase core (dimerization and catalytic domains) of an HPK9, the Thermotoga maritima CheA (Protein Data Bank, PDB, entry 1B3Q) [15]. (b) The catalytic domain of T. maritima CheA complexed with Mg2+ and the nucleotide analog ADPCP (PDB entry 1I58) [16]. (c) The catalytic domain of the HPK2 E. coli EnvZ complexed with ADP (model 1 from PDB entry 1BXD) [14]. (d) The catalytic domain of an HPK3, PhoQ of E. coli, complexed with Mg2+ and the nucleotide analog AMPNP (PDB entry 1ID0) [18]. The figure was created with Swiss-PdbViewer 3.7 and rendered with POV-Ray 3.1.
The sensing and linker domains
On the basis of gene sequence analyses, the typical HPK appears to be a transmembrane sensor. This type of HPK usually has an uncleaved signal sequence, which serves as a first transmembrane helix (TM1), an extracellular sensing domain, and a second transmembrane helix (TM2); this is similar to typical type I tyrosine protein kinase receptors, such as the receptors for the epidermal growth factor (EGF) and insulin [23], but in eukaryotic tyrosine protein kinase receptors the signal sequence is generally cleaved to remove TM1. The extracellular sensing domains of HPKs are extremely diverse and, in contrast to the type I tyrosine protein kinase receptors, no conserved structural motif can be inferred from sequence comparisons [22]. It seems that almost any type of sensory domain can regulate HPK activity. In Gram-negative bacteria some HPKs sense through interaction with a periplasmic binding protein that may also interact with ATP-binding cassette (ABC) transport systems [24].
Inside the cytoplasm, a typical HPK has a HAMP domain, a widely distributed putative regulatory element in transmembrane and other signaling proteins [25], that consists of about 50 residues with two helical segments and a non-helical, compact subdomain in between [25,26]. The HAMP domain (also called the linker) is located between the second transmembrane domain and the dimerization domain (Figure 1) and may play a criticial role in HPK signal transduction [25,26]. It may also directly participate in binding sensory co-factors, such as flavin adenine dinucleotide (FAD) [27]. The HAMP domain might have a structure analogous to that of helix-loop-helix transcription factors [22].
Hybrid kinases
As mentioned above, hybrid HPKs contain both an HPK and an additional carboxy-terminal receiver domain. They are not stand-alone signaling systems, however, but have to communicate with a separate downstream response regulator with an output activity. They achieve this by using a multi-step phosphorelay mechanism, rather than the single phosphotransfer mechanism that operates in normal two-component signaling pathways [28]. In phosphorelays, an intermediate histidine phosphotransfer protein (HPt) is involved either as a soluble protein or as an attached carboxy-terminal domain of the hybrid HPK. HPt proteins receive phosphates from hybrid HPKs and shuttle them to the receiver domains of the downstream response regulators [28]. In certain phosphorelay systems, receiver domains of hybrid HPKs also mediate the hydrolysis of phosphorylated HPt intermediates [29]. The phosphate group is carried on a conserved histidine residue of the HPt protein. A number of three-dimensional structures of HPt domains have been solved, including those of the bacterial HPKs ArcB [30], and CheA [31] as well as the yeast Hpt protein Ypd1 [32]. Despite their diversity of sources, these structures reveal a nearly identical four-helix-bundle core structure [31].
Structures of a range of response regulator receiver domains have also been determined, although not of a hybrid kinase and only of a few intact response regulators. The structure of the receiver domain of the chemotaxis response regulator CheY is generally taken as the stereotypical example of this domain [33,34]. The receiver domain is a five-stranded parallel β sheet that is wound twice, and the site of aspartate phosphorylation is in an acidic pocket near the carboxy-terminal edge of the β sheet [32].
Localization and function
Transmembrane receptor HPKs
Most HPKs are transmembane proteins that are presumed to be receptors for extracellular signals [35]. In the vast majority of cases, the putative ligands are not known. Furthermore, membrane localization does not necessarily mean that the HPK has to bind a soluble extracellular ligand. For example, the well-studied E. coli osmosensing HPK EnvZ is an integral membrane protein that appears to have a periplasmic sensing domain. Removal of this domain does not disrupt osmosensing via EnvZ, however [36], and it is unlikely that EnvZ senses osmolarity by directly binding small soluble molecules. It is more likely that changes in osmolarity modulate interactions of EnvZ with outer membrane proteins, or affect membrane tension, and such changes might in some other way shift the equilibrium between kinase and phosphatase activity of EnvZ. A similar mechanism might regulate the activity of the osmosensing HPK Sln1p in the yeast Saccharomyces cerevisiae, given that the Sln1p transmembrane regions are required for transducing changes in osmolarity [37].
Soluble HPKs
Some HPKs are soluble cytosolic proteins that couple to discrete transmembrane receptors, and another group of HPKs are predicted to be soluble proteins that so far have no known membrane-linked receptor. The best-studied example of a soluble HPK that couples to transmembrane receptors is CheA [11,22]. In E. coli, CheA couples to five different chemotaxis receptors, four of which bind - either directly or through the use of a separate binding protein - small ligands, such as amino acids and sugars, which in turn act as chemoattractants for the bacteria. Through interactions between the receptors and CheA, ligand binding regulates CheA HPK activity and thus controls activity of the chemotaxis two-component system. This situation is operationally very similar to that of mammalian cytokine receptors that control the activity of soluble cytosolic tyrosine protein kinases: like the E. coli chemotaxis receptors, cytokine receptors have no enzymatic activity of their own, but couple to intracellular protein kinases [23,38].
In a similar vein, receptor HPKs function analogously to growth factor receptors with tyrosine kinase activity in animal cells: both classes of receptor are type 1 membrane proteins with an amino-terminal extracellular sensing domain that is linked to the intracellular signaling domain via a single transmembrane helix. Both types of receptor also function as dimers, trans-phosphorylating their partner subunit [23]. But whereas phosphotyrosine-bearing receptors act as binding sites for Src-homology 2 (SH2) domain proteins, autophosphorylated receptor HPKs transfer their phosphate to downstream response regulators, and so must in fact become dephosphorylated in order to transmit their signals. It is interesting to note that there are currently no examples of organisms that have both receptor tyrosine kinases and receptor HPKs, and this suggests that these receptors fulfill equivalent roles.
The E. coli chemotaxis receptors that regulate CheA show higher-order organization in the bacterial inner membrane, where they are predominantly clustered in one macromolecular patch [39]. Although the chemotaxis receptors themselves are not HPKs, they might provide a model for the organization of those HPKs that function as receptors. The higher-order structures are thought to be necessary for the exquisite sensitivity and dynamic signaling properties of the chemotaxis signaling system [40]. Although chemotaxis may be a specialized signaling system, in principle its properties could be important for other signaling pathways that must be able to respond to small changes in ligand concentration and/or over a wide range of ligand concentrations.
Mechanism
The responses generated by HPK signaling are mediated by response regulators, which - like HPKs - form a large family of proteins [1]. Most bacterial response regulators have an amino-terminal receiver domain containing the site of aspartate phosphorylation. Signaling proceeds by autophosphorylation of the HPK domain followed by phosphotransfer to a receiver domain [41,42]. A carboxy-terminal DNA-binding domain allows the response regulator to function as a transcriptional regulator [28]. The presence of the conserved receiver domain defines a protein as a response regulator, but not all response regulators are transcription factors. Other examples of output activities include enzymatic activity, such as that of the receptor methylesterase CheB [28], responsible for adaptation in bacterial chemotaxis, and protein-protein interaction, mediated, for example, by the chemotaxis effector protein CheY [28]. Regardless of the particular output function of a response regulator, its activity is strongly dependent on the state of aspartate phosphorylation of its receiver domain. Thus, extracellular signals are transduced into a cytosolic output response via a protein phosphorylation cascade mediated by an HPK and a response regulator.
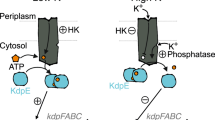
HPK sensor domains regulate kinase activity, but as discussed above, many HPKs also have a phosphatase activity [3,13]. When the kinase state predominates, the net activity of the two-component pathway is phosphorylation of the response regulator; when the phosphatase state predominates, the net activity is dephosphorylation of the response regulator. Because two antagonistic states are in equilibrium, ligands exert tight regulation over pathway activity because a shift in the equilibrium between kinase and phosphatase steady states is amplified by zero-order ultrasensitive kinetic effects [43,44].
In many HPKs, the regulatory effects of the phosphatase activity predominate [3,45]. Thus, the dominant phenotype of a mutant strain lacking a particular receptor-HPK is often a low-level constitutive activity of the response regulator under conditions in which, in wild-type cells, phosphatase activity of the HPK leads to response regulator inactivation. Regulation of these dual-function receptor-HPKs appears to involve modulation of a balance between two distinct states, namely kinase on and phosphatase off, or kinase off and phosphatase on [46]. This phosphatase activity is not simply a reverse phosphotransfer, however, because phosphatase activity is retained in some mutant HPKs lacking the H-box histidine [47], and most HPKs do not have intrinsic autophosphatase activity.
Frontiers
HPKs and two-component systems are an exciting area of research for a growing number of research groups studying the basic biochemistry of these systems as well as those focused on medically relevant topics such as pathogenesis, quorum sensing, and biofilm formation. Of medical interest is the potential for using HPKs as targets for new classes of antimicrobial drug. HPKs are particularly attractive for this purpose because they are not found in animals, and also because interfering with a pathogen's HPK signaling systems may expose it to destruction by the host immune system rather than being directly toxic. This type of indirect antibiotic would be less likely to evoke resistance in the target strain. Although some potential HPK inhibitors have been discovered [16,48], none of them has yet advanced to the stage of being used as a pharmaceutical. Research in this area is likely to accelerate as more HPK structures become available and can be examined for structure-based drug design.
The HPK10 family is one that needs more basic study to determine structural information. It is a distinct and widely distributed family that includes six-transmembrane HPK receptors with hydrophobic amino-terminal domains; they are referred to as 'six-transmembrane' receptors, but both algorithmic predictions and measurements of topology have determined only that HPK10 receptors have between five and seven transmembrane helices [49,50]. These receptors appear generally to function in cell-cell communication. In the few cases where stimulatory ligands have been identified, they are small peptides or modified peptides that are secreted by the same organism. For example, the AgrC system in Staphylococcus aureus is a quorum-sensing system that exports and then senses modified peptides of eight to nine amino acids that have a thio-lactone ring [51]. There is no direct evidence that the six-transmembrane HPKs are dimers, and the kinase core domains of members of the HPK10 family differ from other HPKs; in particular, the HPK family has no D box, and the region near the H box does not show a high similarity to other HPK dimerization domains [1]. This suggests substantial differences in the geometry of the active site, and transmembrane signal transduction mediated by the HPK10 family may proceed by analogy to the events mediated by eukaryotic seven-transmembrane receptors, whereas signaling through other families of HPK receptors is analogous to signaling through type I tyrosine protein kinases.
Finally, the study of HPK signaling in eukaryotes lags behind that in prokaryotes and many challenges lie ahead, including identifying the signaling connections between hybrid HPKs, HPt proteins and response regulators, and determining the interactions between the phosphorelay pathways and the other signaling pathways in eukaryotic cells. In Arabidopsis thaliana (and presumably in many other plants as well) the HPK gene family has undergone great expansion [5]. In Arabidopsis, at least five different HPK proteins act as receptors for the hormone ethylene, and the genes encoding these receptors show redundancy. Thus, plants with mutations in one, two or even three of these receptor HPKs have no phenotype; only a quadruple mutant strain shows a phenotype, namely a constitutive response to ethylene [6]. This complexity presents a significant practical challenge to the genetic analysis of HPK signaling in plants.
Similar challenges arise in studying Dictyostelium HPK genes, of which there are at least 15 [5]. Rather than responding to the same ligand, it is probable that many of these HPKs signal to the same output pathway (the response regulator RegA that controls intracellular cAMP levels), so the HPKs are likely to have overlapping functions. The HPK signaling pathways that control cAMP levels must be coordinated with other signaling systems, such as G-protein-dependent pathways, that also regulate cAMP levels in the same cells [52]. A similar signal transduction network must also operate in other eukaryotic HPK systems that are known to control the activity of mitogen-activated protein (MAP) kinase pathways, such as those involved in ethylene signaling in Arabidoposis and osmosensing in yeast [6,42]. Our traditional view of signal transduction is heavily biased towards the mechanisms used by animal cells, and so HPKs are not the main focus of studies of signal-transduction enzymes, but outside the animal kingdom they are the most important transducers of extracellular signals.
References
Grebe TW, Stock JB: The histidine protein kinase superfamily. Adv Microb Physiol. 1999, 41: 139-227. The first thorough classification of HPK and response regulator families on the basis of sequence alignments and motifs.
Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR: Evolution of two-component signal transduction. Mol Biol Evol. 2000, 17: 1956-1970. An attempt to classify HPKs and response regulators into families, which includes some interesting speculation on evolutionary relationships between HPKs and eukaryotic protein kinases.
Hoch JA, Silhavy TJ: Two-component signal transduction. 1995, Washington, D.C.: ASM Press, A collection of reviews by experts in two-component signal transduction that is considered the starting point for most literature searches. This is still the best source of reviews for some areas, though others have become dated.
Fuqua C, Parsek MR, Greenberg EP: Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001, 35: 439-468. 10.1146/annurev.genet.35.102401.090913. A review of work on quorum sensing via acylated homoserine lactones, including their production and their detection by receptor HPKs.
Thomason P, Kay R: Eukaryotic signal transduction via histidine-aspartate phosphorelay. J Cell Sci. 2000, 113: 3141-3150. A review of HPK systems in eukaryotes and their interaction with downstream pathways such as MAP kinase cascades and the cAMP/PKA pathway.
Hua J, Meyerowitz EM: Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998, 94: 261-271. An outstanding genetic study of ethylene receptor signaling in plants. Isolation of loss-of-function alleles of four ethylene receptor genes shows functional redundancy between the receptors, and also demonstrates that ethylene acts to inhibit the receptors rather than activating them.
The C. elegans Sequencing Consortium: Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998, 282: 2012-2018. 10.1126/science.282.5396.2012. Sequencing of the 97-megabase C. elegans genome was completed in 1998 and was the first genome of a multicellular organism to be fully sequenced.
Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al: The genome sequence of Drosophila melanogaster. Science. 2000, 287: 2185-2195. 10.1126/science.287.5461.2185. This paper reports the completion of the genome sequence of D. melanogaster, one of the most important experimental models in biology.
International Human Genome Sequencing Consortium: Initial sequencing and analysis of the human genome. Nature. 2001, 409: 860-921. 10.1038/35057062. A draft sequence of the human genome and intial analysis of the data.
Matsubara M, Kitaoka SI, Takeda SI, Mizuno T: Tuning of the porin expression under anaerobic growth conditions by His-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes Cells. 2000, 5: 555-569. 10.1046/j.1365-2443.2000.00347.x. An excellent example of networking between two-component signal transduction systems.
Stock JB, Surette M: Chemotaxis. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Edited by: Neidhardt FC. 1996, Washington, DC: ASM, 1103-1129. A detailed and thorough review of the chemotaxis system in E. coli and Salmonella, though some sections have been superseded by more recent research.
Dutta R, Qin L, Inouye M: Histidine kinases: diversity of domain organization. Mol Microbiol. 1999, 34: 633-640. 10.1046/j.1365-2958.1999.01646.x. A review and hypothesis paper that includes some speculations about HPK architecture and the interrelation between the different HPK domains.
Perego M, Hoch JA: Protein aspartate phosphatases control the output of two-component signal transduction systems. Trends Genet. 1996, 12: 97-101. 10.1016/0168-9525(96)81420-X. A review of the role of HPK phosphatase activity and HPK-independent phosphatases in controlling the output of two-component systems.
Tanaka T, Saha SK, Tomomori C, Ishima R, Liu D, Tong KI, Park H, Dutta R, Qin L, Swindells MB, et al: NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature. 1998, 396: 88-92. 10.1038/23968. The first structural analysis of an HPK catalytic domain and demonstration of the HPK structural homology to Hsp90 and DNA gyrase B.
Bilwes AM, Alex LA, Crane BR, Simon MI: Structure of CheA, a signal-transducing histidine kinase. Cell. 1999, 96: 131-141. The first X-ray crystal structure of an HPK, and the only one that includes both the dimerization and catalytic domains.
Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI: Nucleotide binding by the histidine kinase CheA. Nat Struct Biol. 2001, 8: 353-360. 10.1038/86243. An important paper that shows the structural effects of binding different nucleotide analogs to the CheA catalytic domain.
Ban C, Junop M, Yang W: Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999, 97: 85-97. A seminal paper that provides a basis for understanding the relation of ATPase activity to conformational transitions in MutL and its homologs, such as the HPKs.
Marina A, Mott C, Auyzenberg A, Hendrickson WA, Waldburger CD: Structural and mutational analysis of the PhoQ histidine kinase catalytic domain: insight into the reaction mechanism. J Biol Chem. 2001, 276: 41182-41190. 10.1074/jbc.M106080200. This paper presents a high-resolution X-ray structure of the PhoQ catalytic domain with an ATP analog bound. Together with the papers by Bilwes et al. [15,16], it provides a good understanding of the HPK catalytic domain structure.
Stock A, Chen T, Welsh D, Stock J: CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci USA. 1988, 85: 1403-1407. The first sequence of a CheA protein and analysis that shows its relatedness to other HPKs through conserved sequence motifs.
Parkinson JS, Kofoid EC: Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992, 26: 71-112. 10.1146/annurev.ge.26.120192.000443. A review of two-component systems in which the receiver domain and transmitter domain terminology was introduced, as well as the designation of various HPK sequence motifs as
Tomomori C, Tanaka T, Dutta R, Park H, Saha SK, Zhu Y, Ishima R, Liu D, Tong KI, Kurokawa H, et al: Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999, 6: 729-734. 10.1038/11495. The first structure of the dimerization domain of an orthodox HPK, which shows a close relationship to the CheA structure.
Wolanin PW, Stock JB: Transmembrane signaling and the regulation of histidine kinase activity. In Histidine Kinases in Signal Transduction. Edited by: Inouye M, Dutta R. New York: Academic Press, A forthcoming thorough review of HPK signaling in a new collection of reviews on two-component systems.,
Schlessinger J: Cell signaling by receptor tyrosine kinases. Cell. 2000, 103: 211-225. The most recent and thorough review of receptor tyrosine kinase signaling.
Quiocho FA, Ledvina PS: Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996, 20: 17-25. A review of periplasmic binding protein structures and their roles in transport and chemotaxis.
Aravind L, Ponting CP: The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999, 176: 111-116. 10.1016/S0378-1097(99)00197-4. Definition of the HAMP domain family as a widely occurring element in signal transduction systems and identification of the HPK 'linker as an important representative of this family.
Williams SB, Stewart V: Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol Microbiol. 1999, 33: 1093-1102. 10.1046/j.1365-2958.1999.01562.x. An examination of, and speculation on the role of, the linker region (or HAMP domain) in HPK transmembrane signaling.
Bibikov SI, Barnes LA, Gitin Y, Parkinson JS: Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc Natl Acad Sci USA. 2000, 97: 5830-5835. 10.1073/pnas.100118697. This paper demonstrates conclusively that Aer binds FAD and explores the relative roles of the PAS (a widely distributed sensory domain that is often involved in binding prosthetic groups such as heme or FAD) and HAMP domains in FAD binding.
Stock AM, Robinson VL, Goudreau PN: Two-component signal transduction. Annu Rev Biochem. 2000, 69: 183-215. 10.1146/annurev.biochem.69.1.183. A recent and comprehensive review of two-component signaling that focuses especially on protein structures.
Freeman JA, Lilley BN, Bassler BL: A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 2000, 35: 139-149. 10.1046/j.1365-2958.2000.01684.x. A detailed analysis of a phosphorelay network in bacteria that controls quorum sensing.
Kato M, Shimizu T, Mizuno T, Hakoshima T: Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor protein ArcB complexed with the chemotaxis response regulator CheY. Acta Crystallogr D Biol Crystallogr. 1999, 55: 1257-1263. 10.1107/S0907444999005053. This is the second X-ray structure of the ArcB HPt domain, and the only structure of an HPt domain in complex with a response regulator.
Mourey L, Da Re S, Pédelacq J-D, Tolstykh T, Faurie C, Guillet V, Stock JB, Samama J-P: Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J Biol Chem. 2001, 276: 31074-31082. 10.1074/jbc.M101943200. The first X-ray crystal structure of the HPt domain of the CheA family of HPKs. The paper includes a detailed comparison with the structure of other HPt proteins, and argues against the conclusions advanced by Appleby et al. [41].
Xu Q, West AH: Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J Mol Biol. 1999, 292: 1039-1050. 10.1006/jmbi.1999.3143. The first structure of the yeast HPt protein Ypd1p and an analysis suggesting that HPt proteins have a common, distinct core structure.
Stock AM, Mottonen JM, Stock JB, Schutt CE: Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989, 337: 745-749. 10.1038/337745a0. The first X-ray crystal structure of a response regulator.
Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE: Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem. 2001, 276: 16425-16431. 10.1074/jbc.M101002200. The first crystal structure of CheY complexed with Mg2+ and BeF3-, a phosphate analog, and a comparison of this structure with other CheY structures.
Hoch JA: Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000, 3: 165-170. 10.1016/S1369-5274(00)00070-9. A recent review by the original discoverer of phosphorelays. Hoch points out that we should not be tempted to use the few well-characterized two-component and/or phosphorelay systems so far studied as a universal template for the function of all the others. We are also a long way from truly understanding the function of any single pathway.
Leonardo MR, Forst S: Re-examination of the role of the periplasmic domain of EnvZ in sensing of osmolarity signals in Escherichia coli. Mol Microbiol. 1996, 22: 405-413. 10.1046/j.1365-2958.1996.1271487.x. This paper reports the surprising finding that the periplasmic domain of the HPK EnvZ could be completely replaced without disruption of osmosensing, suggesting that EnvZ senses by a mechanism other than ligand binding.
Ostrander DB, Gorman JA: The extracellular domain of the Saccharomyces cerevisiae Sln1p membrane osmolarity sensor is necessary for kinase activity. J Bacteriol. 1999, 181: 2527-2534. The authors presents evidence that the extracellular domain of Sln1p may be a dimerization region required for Sln1p kinase activity. The first transmembrane domain of Sln1p is also required for sensing osmolarity.
Taniguchi T: Cytokine signaling through nonreceptor protein tyrosine kinases. Science. 1995, 268: 251-255. A review of the functions of cytokine receptors and their interactions with soluble protein tyrosine kinases.
Maddock JR, Shapiro L: Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993, 259: 1717-1723. An important paper reporting the discovery that chemotaxis receptors are localized in clusters at the cell pole.
Stock J, Levit M: Signal transduction: hair brains in bacterial chemotaxis. Curr Biol. 2000, 10: R11-14. 10.1016/S0960-9822(99)00248-1. A minireview advancing the idea that receptor clusters, such as those discovered by Maddock and Shapiro [39], have much greater information-processing capacity than previously appreciated.
Appleby JL, Parkinson JS, Bourret RB: Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996, 86: 845-848. An influential but speculative minireview, written to accompany Posas et al. [42], which posits the existence of phosphorelays as a widespread and general biochemical mechanism on the basis of superficial similarities in protein structure and function.
Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H: Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 'two-component' osmosensor. Cell. 1996, 86: 865-875. Discovery of the first eukaryotic phosphorelay. The Ypd1 gene encodes an HPt protein that transfers phosphate between the hybrid HPK Sln1p and the response regulator Ssk1p.
Koshland DE, Goldbeter A, Stock JB: Amplification and adaptation in regulatory and sensory systems. Science. 1982, 217: 220-225. This seminal paper defined the relationship between gain and sensitivity in biological systems, and introduced the concept of ultrasensitivity.
Russo FD, Silhavy TJ: The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1993, 1: 306-310. 10.1016/0966-842X(93)90007-E. A study of the ultrasensitivity that is achieved in the EnvZ-OmpR system through the balance of kinase and phosphatase activities.
Zhu Y, Qin L, Yoshida T, Inouye M: Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci USA. 2000, 97: 7808-7813. 10.1073/pnas.97.14.7808. Definitive experimental results showing that the dimerization domain of HPKs can retain response regulator phosphatase activity in the absence of the catalytic domain.
Hsing W, Russo FD, Bernd KK, Silhavy TJ: Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998, 180: 4538-4546. A mutational analysis showing the distinction between kinase and phosphatase activities in terms of both structure and function.
Hsing W, Silhavy TJ: Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997, 179: 3729-3735. A demonstration that the phosphorylated histidine is important, but not essential, for HPK phosphatase activity.
Macielag MJ, Goldschmidt R: Inhibitors of bacterial two-component signalling systems. Expert Opin Investig Drugs. 2000, 9: 2351-2369. A review of past attempts to find specific inhibitors of two-component signaling and of more recent efforts using structure-based drug design.
Piazza F, Tortosa P, Dubnau D: Mutational analysis and membrane topology of ComP, a quorum-sensing histidine kinase of Bacillus subtilis controlling competence development. J Bacteriol. 1999, 181: 4540-4548. One of the few experimental analyses of the membrane topology of a six-transmembrane HPK receptor, ComP, using PhoA and LacZ fusions as well as proteolytic studies.
Ethier J, Boyd JM: Topological analysis and role of the transmembrane domain in polar targeting of PilS, a Pseudomonas aeruginosa sensor kinase. Mol Microbiol. 2000, 38: 891-903. 10.1046/j.1365-2958.2000.02189.x. Another of the rare experimental studies of the membrane topology of a six-transmembrane HPK receptor using phoA and lacZ fusions.
Lyon GJ, Mayville P, Muir TW, Novick RP: Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc Natl Acad Sci USA. 2000, 97: 13330-13335. 10.1073/pnas.97.24.13330. This paper describes the rational design of a molecule that inhibits virulence in Staphylococcus aureus through interaction with an HPK receptor.
Thomason PA, Traynor D, Stock JB, Kay RR: The RdeA-RegA system, a eukaryotic phosphorelay controlling cAMP breakdown. J Biol Chem. 1999, 274: 27379-27384. 10.1074/jbc.274.39.27379. The first biochemical demonstration of a phosphorelay between the Dictyostelium proteins RdeA and RegA, and the activation of RegA by aspartate phosphorylation.
Acknowledgements
P.M.W. was supported by NIH fellowship 1F32 GM064228. P.A.T. was supported by a fellowship from the Wellcome Trust. This work was also supported by a grant from the Defense Advanced Research Projects Agency (3416183).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wolanin, P.M., Thomason, P.A. & Stock, J.B. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 3, reviews3013.1 (2002). https://doi.org/10.1186/gb-2002-3-10-reviews3013
Published:
DOI: https://doi.org/10.1186/gb-2002-3-10-reviews3013