Abstract
ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) genes are a family of plant specific transcription factors, which play an important role in the regulation of plant lateral organ development and metabolism. However, a genome-wide analysis of the AS2/LOB gene family is still not available for barley. In the present study, 24 AS2-like (ASL)/LOB domain (LBD) genes were identified based on the barley (Hordeum vulgare L.) genome sequence. A phylogenetic tree of ASL/LBD proteins from barley, Arabidopsis, maize, and rice was constructed. The ASL/LBD genes were classified into two classes, class I and class II, which were divided into five and two subgroups, respectively. Genes homologous in barley and Arabidopsis were analyzed. In addition, the structure and chromosomal locations of the genes were analyzed. Expression profiles indicated that barley HvASL/LBD genes exhibit a variety of expression patterns, suggesting that they are involved in various aspects of physiological and developmental processes. This genome-wide analysis of the barley AS2/LOB gene family contributes to our understanding of the functions of the AS2/LOB gene family.
中文概要
目 的
从大麦全基因组中鉴定AS2/LOB 基因家族,并进行基因进化、基因结构、染色体定位以及组织、表达分析,为大麦ASL/LBD 基因进一步功能研究与鉴定奠定基础。
创新点
首次在大麦全基因组水平上分析AS2/LOB 基因家族,并对部分基因的功能进行预测和分析。
方 法
利用大麦基因组数据库,通过生物信息学手段,鉴定大麦AS2/LOB 基因家族成员;采用MEGA6软件进行系统进化树分析; 利用GSDS 及MapDraw 工具进行基因结构及染色体定位分析;利用已有的大麦RNAseq 数据进行组织表达谱分析。
结 论
通过全基因组分析,大麦AS2/LOB 家族基因包括24 个成员,在进化上分为两大类,7 个亚家族,分布于大麦7 条染色体上,组织表达模式具有多样性,与已经报道的ASL/LBD 基因具有良好的同源性。这些信息为大麦AS2/LOB 基因家族的功能分析奠定了基础。
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The plant-specific ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family contains transcription factors which play an important role in the regulation of plant lateral organ development. AS2-like (ASL)/LOB domain (LBD) genes are also involved in the regulation of anthocyanin and nitrogen metabolism (Rubin et al., 2009; Majer and Hochholdinger, 2011; Koppolu et al., 2013; Xu et al., 2016). ASL/LBD genes encode a protein containing a conserved amino acid domain of unknown function, termed the AS2/LOB domain. AS2/LOB domain recognized a 6-bp GCGGCG consensus motif and interacts with a specific basic helix-loop-helix (bHLH) protein (Husbands et al., 2007). According to the structure of the AS2/LOB domain in the N-terminus, the AS2/LOB gene family can be divided into two classes, class I and class II. Class I contains a conserved CX2CX6CX3C zinc finger-like motif and an LX6LX3LX6L leucine zipper-like coiled-coil motif (Shuai et al., 2002). There are also two conserved blocks, the C block and GAS block, in the AS2/LOB domain of the class I proteins (Husbands et al., 2007). However, class II ASL/LBD genes have only a conserved zinc fingerlike domain (Shuai et al., 2002).
In Arabidopsis, 43 AS2/LOB gene family members have been identified (Shuai et al., 2002; Matsumura et al., 2009). Based on a genome scan of the published genome sequence and protein sequence similarity in different species, 35 rice and 44 maize ASL/LBD genes have been reported. Expression analysis suggests that these genes are transcribed in a wide variety of tissues and organs (Yang et al., 2006; Zhang et al., 2014). Lateral organs of a higher plant are initiated from small cell groups on the flanks of the dome-shaped shoot apical meristem (SAM) (Borghi et al., 2007). Maintenance of an active shoot meristem requires expression of homeobox KNOX genes, such as SHOOT MERISTEMLESS (STM) of Arabidopsis, which are excluded from organ primordia (Long et al., 1996). AS1 is expressed in organ initials and physically interacts with AS2, which encodes nuclear protein with the plant-specific AS2/LOB domain, to repress KNOX gene expression, thus guiding primordia towards differentiation (Ori et al., 2000; Semiarti et al., 2001; Byrne et al., 2002; Guo et al., 2008). The AS1-AS2 complex also represses the ETTIN gene directly and the ETTIN and Auxin Response Factor 4 (ARF4) genes indirectly through trans-acting short-interfering RNA (tasiR)-ARF in adaxial-abaxial specification of Arabidopsis leaves (Iwasaki et al., 2013). AS2 is also expressed in the adaxial parts of leaf primordia and young floral organs (Iwakawa et al., 2007; Keta et al., 2012). In cooperation with AS1 and JAGGED (JAG), it restricts the boundary cells in the floral organs. Genetic analysis showed that AS1, AS2, and JAG genes function in the sepal and petal primordia to repress boundary-specifying genes (CUC1, CUC2, and PETAL LOSS) to promote normal development of the organs (Xu et al., 2008). Normal maize ears are unbranched and tassels have long branches only at their base. However, the ramosa2 (ra2) mutant of maize results in increased branching, with short branches replaced by long indeterminate ones (Bortiri et al., 2006). Function analysis showed that ra2 encodes the AS2/LOB domain transcription factor which determines the fate of stem cells in branch meristems of maize (Bortiri et al., 2006). Similarly, Vrs4 is the ortholog of maize RAMOSA2 in barley. Genetic mapping and mutant analysis revealed that Vrs4 controls spikelet determinacy and row-type in barley (Koppolu et al., 2013).
Plant root development is also affected by ASL/LBD genes, including lateral root development in Arabidopsis, rice, and maize (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007; Taramino et al., 2007). AtARF7 and AtARF19 regulate lateral root formation as transcriptional activators of early auxin response genes in Arabidopsis thaliana (Okushima et al., 2007). Further analysis revealed that AtARF7 and AtARF19 directly regulate the auxin-mediated transcription of AtASL18/LBD16 and AtASL16/LBD29 in roots (Okushima et al., 2007). Overexpression of AtASL18/LBD16 and AtASL16/LBD29 induces lateral root formation in the absence of AtARF7 and AtARF19. In addition, AASL20/LBD18 in conjunction with AtASL18/LBD16 functions in the initiation and emergence of lateral roots as a downstream regulator of AtARF7 and AtARF19 (Lee et al., 2009). These data suggest that ASL/LBD genes mediate lateral root formation in Arabidopsis by different molecular pathways. In maize, conventional genetic approaches have led to the identification of ASL/LBD genes that function in plant growth and development. For example, the rtcs (rootless concerning crown and seminal roots, Zmlbd2) mutant is impaired in the initiation of the embryonic seminal roots and the post-embryonic shoot-borne root system (Taramino et al., 2007). The RTCL (RTCS-like, ZmLBD43) gene is a paralog of RTCS, which displays spatio-temporal expression patterns in roots that are highly correlated with those of the RTCS gene (Taramino et al., 2007). Both RTCS and RTCL proteins are auxin-responsive genes involved in the early events that lead to the initiation and maintenance of seminal and shoot-borne root primordia formation. Both act as transcription factors and bind to downstream promoters of ASL/LBD genes (Taramino et al., 2007). Taken together, these results suggest that ASL/LBD genes function in lateral and seminal root initiation and emergence, as well as shoot-borne root primordia formation.
To date, only a subset of the AS2/LOB gene family has been systematically analyzed based on genome sequencing databases (Iwakawa et al., 2002; Shuai et al., 2002; Yang et al., 2006, Wang et al., 2013; Zhang et al., 2014). Several ASL/LBD genes associated with mutant phenotypes involving many aspects of plant development, including embryo, root, leaf, and inflorescence development, have been functionally characterized (Majer and Hochholdinger, 2011). Therefore, a major focus of this study was to gain a better understanding of the barley AS2/LOB gene family and its expression pattern in different tissues. Barley is one of the world’s earliest domesticated and most important plant crops. It has been used as animal fodder, as a source of fermentable material for beer, and as a component of various health foods. However, the barley AS2/LOB gene family has not been characterized in detail. Recently, physical, genetic, and functional sequence maps of the barley genome have been published which can be used to screen the AS2/LOB gene family (International Barley Genome Sequencing Consortium et al., 2012). In the present study, we provide detailed information on the genomic structure, chromosomal locations, sequence homology, and expression patterns of barley ASL/LBD genes. A phylogenetic tree of ASL/LBD genes in barley, Arabidopsis, maize, and rice was also constructed, which will help future studies aimed at elucidating the vital roles of HvASL/LBD genes in barley developmental processes.
2 Materials and methods
2.1 Sequence database searches
Multiple database searches were performed to collect all members of the barley AS2/LOB gene members. Barley sequence data were sourced from the Morex assembly (International Barley Genome Sequencing Consortium et al., 2012) and National Center for Biotechnology Information (NCBI) database. We used the BLAST programs (TBLASTN and BLASTN) available on the Institute of Plant Genetics and Crop Plant Research (IPK) barley genome database and NCBI barley expressed sequence tag (EST) database. As a query sequence, we used the amino acid sequence of the AS2/LOB domain from Arabidopsis thaliana, rice (Oryza sativa L.), and maize (Zea mays L.) ASL/LBD genes (http://planttfdb.cbi.pku.edu.cn). To increase the extent of the database search results, we also performed database searches using amino acid sequences of some members of the barley AS2/LOB gene family as query sequences to confirm completion of the collection. All hits with expected (E) values less than 1.0 were retrieved and the non-redundant sequences were examined for the presence of the conserved AS2/LOB domain using domain analysis programs Pfam (protein family; http://pfam.sanger.ac.uk) and SMART (simple modular architecture research tool; http://smart.embl-heidelberg.de) with the default cutoff parameters (Letunic et al., 2012; Punta et al., 2012). All the ASL/LBD proteins contained full length sequence.
Isoelectric points and protein molecular weights were obtained with the help of the proteomics and sequence analysis tools on the ExPASy proteomics server (http://expasy.org) (Artimo et al., 2012). Putative promoter sequences (2000 bp upstream from the 5′ UTR region) of HvASL/LBD genes were obtained from the draft barley genome sequence (International Barley Genome Sequencing Consortium et al., 2012) and a search for hormone responsive elements was performed using the PlantCARE database (Lescot et al., 2002).
2.2 Chromosomal location and structure of the HvASL/LBD genes
Chromosomal locations and genomic sequences were retrieved from the barley genome database that was downloaded from the IPK database. All genes were mapped to the chromosomes with MapDraw software (Liu and Meng, 2003). The exon/intron structures were constructed using GSDS (gene structure display server; http://gsds.cbi.pku.edu.cn) (Hu et al., 2015). The HvASL/LBD proteins were named sequentially according to AS2/LOB domain blast results (Iwakawa et al., 2002) and their placement in the barley chromosomes, respectively.
2.3 Sequence analysis and construction of the phylogenetic tree
Full-length amino acid sequences of ASL/LBD genes identified in Arabidopsis, maize, rice, and barley were aligned using the Clustal X 1.83 program with default pairwise and multiple alignment parameters (Husbands et al., 2007). The phylogenetic tree was constructed based on this alignment result using the neighbor joining (NJ) method in MEGA Version 6 (Tamura et al., 2013) with the following parameters: Poisson correction, pairwise deletion, uniform rates, and bootstrap (1000 replicates). Conserved motifs were investigated by multiple alignment analyses using MEME Version 3.0 (Bailey and Elkan, 1994).
2.4 Expression analysis of the HvASL/LBD genes
Gene expression data from the cultivar “Morex” were obtained by making use of the barley genome database (http://apex.ipk-gatersleben.de/apex/f?p=284:10:6281639160219::NO). Eight tissues of “Morex” (three biological replications each) earmarking sequential stages of the barley life cycle were selected for deep RNA sequencing (RNA-seq) (International Barley Genome Sequencing Consortium et al., 2012). The tissues comprised: 4-d embryos dissected from germinating grains, roots, and shoots from seedlings (10 cm shoot stage), young developing inflorescences (5 mm), developing inflorescences (1.0–1.5 cm), developing tillers at the six-leaf stage (the third internode), and developing grains at 5 and 15 d post-anthesis (DPA) (bracts removed). The expression patterns are presented as heat maps in green/yellow/red coding, which reflected the fragments per kb of transcript per million mapped reads (FPKM), with red indicating high, yellow medium, and green low expression levels.
3 Results
3.1 Identification of the AS2/LOB family genes in barley
To identify the AS2/LOB gene family members in barley, BLAST searches of the barley databases were performed using the AS2/LOB domain (DUF260 domain) of the Arabidopsis, maize, and rice proteins as a query sequence, and then SMART and Pfam tools were used to check the domains. Twenty-four genes were identified as possibly encoding the AS2/LOB domain (Tables 1 and S1). There were 4, 2, 5, 10, 1, 1, and 1 genes located on chromosomes 1H to 7H (Table 1), respectively. The gene identifier, chromosome position, length of coding sequence, length of amino acid sequence, molecular weight, and pI (isoelectric point) are detailed in Table 1. The identified HvASL/LBD proteins had from 177 (HvASL10/LBD22) to 378 (HvASL7/LBD10) amino acids, a protein mass from 18.89 kD (HvASL11/LBD13) to 40.99 kD (HvASL7/LBD10) and protein pIs ranging from 4.55 (HvASL7/LBD10) to 10.46 (HvASL19/LBD17).
3.2 Phylogenetic and gene structure analysis of the ASL/LBD genes
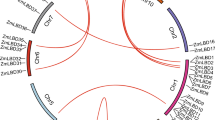
To clarify the phylogenetic relationships among the 24 barley ASL/LBD proteins, alignment analyses were performed using full length amino acid sequences of the HvASL/LBD proteins. The alignment indicated that the HvASL/LBD proteins were identified as two monophyletic subfamilies (class I and class II) (Fig. 1), including 19 and 5 HvASL/LBD genes, respectively, and 5 sister pairs of paralogous ASL/LBD genes (HvASL1/LBD1 and HvASL10/LBD22, HvASL2/LBD2 and HvASL6/LBD9, HvASL3/LBD3 and HvASL8/LBD11, HvASL4/LBD4 and HvASL9/LBD16, HvASL15/LBD21 and HvASL17/LBD24). Sister pairs of paralogous ASL/LBD genes (10/24, 41.67%) had high bootstrap support (bootstrap >90%).
Structural analyses provide valuable information concerning duplication events when interpreting phylogenetic relationships within gene families. Thus, we analyzed the exon/intron structures of the AS2/LOB family genes (right panel in Fig. 1). In barley, the number of exons ranged from 1 within 10 genes (HvASL4/LBD4, HvASL20/LBD6, HvASL22/LBD7, HvASL5/LBD8, HvASL7/LBD10, HvASL18/LBD15, HvASL13/LBD18, HvASL14/LBD19, HvASL24/LBD20 and HvASL10/LBD22) to 5 within a single gene (HvASL19/LBD17). The remaining HvASL/LBD genes had 2 (HvASL1/LBD1, HvASL2/LBD2, HvASL3/LBD3, HvASL21/LBD5, HvASL6/LBD9, HvASL12/LBD12, HvASL11/LBD13, HvASL23/LBD14, HvASL9/LBD16, HvASL15/LBD21, HvASL16/LBD23 and HvASL17/LBD24) or 3 (HvASL8/LBD11) exons. Some members within the same subgroup shared a similar intron/exon structure and gene length (HvASL2/LBD2 and HvASL6/LBD9, HvLBD21/24 and HvLBD6/7/20). The conserved intron/exon structure in each subgroup supported their close evolutionary relationship and the stated classification of subfamilies.
To compare the evolutionary patterns of barley HvASL/LBD proteins with those of other plants and then group them into established subfamilies, a phylogenetic tree was generated using Arabidopsis, maize, rice, and barley full length protein sequences (Fig. 2). The tree suggests two major classes of ASL/LBD genes, characterized by the presence (class I) or absence (class II) of functional leucine-zipper-like domains (Shuai et al., 2002). Class I and class II were subdivided into 5 and 2 groups, respectively. In class I, the subgroups Ia to Ie included 45, 9, 37, 23, and 7 genes, respectively, while the class II group comprised 11 genes in subgroup IIa and 14 genes in subgroup IIb. Subgroups Ia and Ic were the largest (45 and 37 ASL/LBD genes, respectively), and included 9 and 6 barley ASL/LBD genes, respectively. Subgroups Ib, Id, and Ie contained 2, 1, and 1 barley ASL/LBD genes, respectively. In addition, there were 11 ASL/LBD genes in subgroup IIa, of which 2 were from barley, and 14 in subgroup IIb, of which 3 were from barley (Fig. 2). Interestingly, most of the Arabidopsis (dicot) ASL/LBD proteins clustered separately from those of maize, rice, and barley (monocots) (Fig. 2). For example, AtASL39/LBD37, AtASL40/LBD38, and AtASL41/LBD39 clustered independently from ASL/LBD genes in barley (HvASL21/LBD5 and HvASL23/14), maize (ZmLBD7, ZmLBD13, and ZmLBD14) (Zhang et al., 2014), and rice (Os07g40000.1, Os03g33090.1, and Os03g41330.1) (Yang et al., 2006), indicating an evolutionary dichotomy of ASL/LBD genes between dicot and monocot plants.
Phylogenetic analysis of ASL/LBD genes in Arabidopsis, maize, rice, and barley
The gene names used for AtASL/LBDs, ZmLBDs were according to Matsumura et al. (2009) and Zhang et al. (2014); the rice gene ID was derived from Oryza sativa subsp. indica (Yang et al., 2006). AtLBD34 does not contain a complete AS2/LOB domain and was therefore omitted from the phylogenetic tree
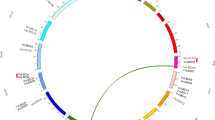
3.3 Chromosomal location analysis of HvASL/LBD genes
Twenty-four genes were located among the 7 barley chromosomes (Table 1, Fig. 3). Chromosome 4 contained the most (10) HvASL/LBD genes. Four genes were identified in 1H and five in 3H. Two HvASL/LBD genes were distributed in 2H, while only one was located on each of chromosomes 5H, 6H, and 7H. Further investigation showed that some members were clustered together within chromosomes (Fig. 3), including HvASL22/LBD7 and HvASL5/LBD8, HvASL23/LBD14, HvASL18/LBD15, HvASL9/LBD16, and HvASL19/LBD17, and HvASL13/LBD18 and HvASL14/LBD19. Precise genetic distances were mapped for a total of 19 genes for the corresponding chromosome. No positional information was found for the genes HvASL3/LBD3, HvASL4/LBD4, HvASL8/LBD11, HvASL14/LBD19, or HvASL24/LBD20.
3.4 Sequence alignment and conserved motifs of HvASL/LBD genes
In Arabidopsis, the ASL/LBD genes have a conserved AS2/LOB domain in the N-terminus of the proteins, and there are two conserved blocks, the C and GAS blocks, in the AS2/LOB domain of the class I proteins. In the present study, sequence alignment results showed that HvASL/LBD genes also contained the C and GAS blocks in the AS2/LOB domains (Fig. 4a). A conserved CX2CX6CX3C zinc finger-like domain was detected within all HvASL/LBD proteins, while an LX6LX3LX6L leucine zipperlike domain was found in only 8 of the class I ASL/LBD genes (Fig. 4b). However, none of the class II proteins were predicted to form coiled-coil structures.
Conserved domains of HvAS2/LOB gene family
(a) AS2/LOB domain element of barley HvASL/LBD proteins. The red dots indicate residues that are conserved in the AS2/LOB domain. (b) The CX2CX6CX3C zinc finger-like domain sequence logos (left) and LX6LX3LX6L leucine zipper-like domain sequence logos (right) (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
3.5 Expression profiles of the HvASL/LBD genes at different developmental stages from RNA-seq data
To investigate the potential functions of the HvASL/LBD genes in barley development, we searched the deep RNA sequencing (RNA-seq) data from eight tissues of the cultivar “Morex” (International Barley Genome Sequencing Consortium et al., 2012). The tissues represent stages of the barley life cycle from germinating grain to the maturing caryopsis. Transcripts from all genes were detected by RNA sequencing except those from the HvASL5/LBD8 gene. The HvASL5/LBD8 gene, also named the Six-rowed spike4 (Vrs4)/HvRAMOSA2 (HvRA2), is an ortholog of the maize inflorescence architecture gene RAMOSA2 (RA2). In situ hybridization analyses revealed that the expression of HvRA2 was first detected during the double ridge stage. At the triple-mound stage, HvRA2 mRNA signals were abundant all over the lateral spikelet primordia with weaker expression in the central spikelet primordia. At the glume primordium stage, HvRA2 mRNAs were detected in both the central and lateral spikelets. Transcript levels of HvRA2 in developing spikes at the triple mound and glume primordium stages were higher than those during later stages (Koppolu et al., 2013). Further analysis revealed that transcripts of eleven genes (HvASL2/LBD2, HvASL21/LBD5, HvASL20/LBD6, HvASL22/LBD7, HvASL6/LBD9, HvASL8/LBD11, HvASL11/LBD13, HvASL23/LBD14, HvASL18/LBD15, HvASL19/LBD17, and HvASL15/LBD21) could be detected in the eight tissues (Fig. 5). Transcripts of HvASL17/LBD24 were detected only in developing grain bracts removed at 5 DPA. Transcripts of HvASL24/LBD20 predominantly present in developing grain bracts removed at 15 DPA, but there was slight expression in roots, young developing inflorescences, and developing tillers (six-leaf and third internode stage) (Fig. 5). These expression patterns could contribute to functional analysis of ASL/LBD genes in barley.
Expression profiles of HvASL/LBD genes (except HvASL5/LBD8 ) in different tissues of barley
Expression patterns are presented as heat maps in green/yellow/red coding, reflecting the fragments per kb of transcript per million mapped reads (FPKM) with red indicating high, yellow medium, and green low expression levels (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
4 Discussion
4.1 Characterization of the barley AS2/LOB gene family
The plant-specific AS2/LOB gene family has a potential role in plant-specific processes (Iwakawa et al., 2002; Shuai et al., 2002; Matsumura et al., 2009; Rubin et al., 2009). Based on plant genome sequencing, 43 ASL/LBD genes have been identified in Arabidopsis (Iwakawa et al., 2002; Shuai et al., 2002; Matsumura et al., 2009), 35 in rice (Yang et al., 2006), and 44 in maize (Zhang et al., 2014). In the present study, 24 HvASL/LBD genes were identified from among 79 379 high- and low-confidence annotated genes listed by the International Barley Genome Sequencing Consortium et al. (2012). Each of them has notable features with a conserved AS2/LOB domain. Remarkably, barley had fewer ASL/LBD genes than Arabidopsis, maize, or rice (Iwakawa et al., 2002; Shuai et al., 2002; Yang et al., 2006; Zhang et al., 2014). This may indicate that other HvASL/LBD genes existing in the unknown genomic regions or chromosome duplication events may have restricted barley evolutionarily expansion.
4.2 Phylogenetic analysis and evolution of barley ASL/LBD genes
Previous phylogenetic analyses have revealed the evolutionary relationships of the ASL/LBD proteins among Arabidopsis, rice, and maize, dividing the AS2/LOB gene family into class I and class II (Iwakawa et al., 2002; Shuai et al., 2002; Yang et al., 2006; Matsumura et al., 2009; Majer and Hochholdinger, 2011; Zhang et al., 2014). In the present study, a phylogenetic tree was constructed using Arabidopsis, rice, maize, and barley ASL/LBD full amino acid sequences. Recently, several ASL/LBD genes associated with mutant phenotypes involved in many aspects of plant development, including embryo, root, leaf, and inflorescence development, have been functionally characterized (Majer and Hochholdinger, 2011). Maize Ra2 encodes an AS2/LOB domain transcription factor, and the ramosa2 (ra2) mutant has increased branching with short branches replaced by long indeterminate ones (Bortiri et al., 2006). In the present study, the barley gene HvASL5/LBD8 (Vrs4/HvRa2) was found to be the ortholog of maize Ra2. Mutant analysis suggested that Vrs4/HvRa2 was a central player in establishing the inflorescence architecture of barley spikes and in determining yield potential and grain number (Koppolu et al., 2013). The ASL/LBD genes CRL1 (ARL1) in rice and RTCS in maize (Fig. 2) are close relatives of AtASL16/LBD29 and are involved in the formation of monocot specific crown roots (Hetz et al., 1996; Inukai et al., 2005; Liu et al., 2005; Taramino et al., 2007). The barley genes HvASL12/LBD12 and HvASL14/LBD19 are close to these genes in the phylogenetic tree (Fig. 2, Table 1), and auxin-responsive elements were also detected in the promoters (Fig. S1). This indicates that HvASL12/LBD12 and HvASL14/LBD19 may be involved in the formation of crown roots in barley. The asymmetric leaves2 (as2) mutant generated abnormal leaves from petioles in a bilaterally asymmetric manner in Arabidopsis (Semiarti et al., 2001), and domain swapping between AS2 and other members of the family showed that the AS2/LOB domain of AS2 was specific for the function of the AS2 gene (Matsumura et al., 2009). In addition, The AS2/LOB domain protein encoded by IG1 (ZmLBD1) is very similar to that of AS2 of Arabidopsis, which also displays abnormal leaf morphology (Evans, 2007). In the present study, HvASL2/LBD2 was the closest orthologous gene of AS2, suggesting that the function of HvASL2/LBD2 should be further investigated. Detecting close phylogenetic relationships and identifying orthologs between monocots and dicots can contribute to the prediction of ASL/LBD gene function in plants (Matsumura et al., 2009; Majer and Hochholdinger, 2011).
4.3 Expression analysis of ASL/LBD genes on plant growth and development
ASL/LBD proteins play a crucial role in defining organ boundaries and are involved in almost all aspects of plant development, including embryo, root, leaf, and inflorescence development (Byrne et al., 2000; Borghi et al., 2007; Majer and Hochholdinger, 2011; Xu et al., 2016). Several Arabidopsis class I members have been implicated in plant development. ASL/LBD gene member AS2 regulates symmetric flat leaf formation by repression of cell proliferation in the adaxial domain (Semiarti et al., 2001; Iwakawa et al., 2007; Iwasaki et al., 2013). Analysis by in situ hybridization showed transcripts of AS2 accumulated in the entire leaf primordium at an early stage (Semiarti et al., 2001; Iwakawa et al., 2007). The AS2 gene was highly expressed in a sample from shoot apices and was involved in leaf adaxial-abaxial polarity (Iwakawa et al., 2002; Iwasaki et al., 2013). In barley, HvASL2/LBD2 was the closest homolog of AS2. It was highly expressed in shoots from seedlings, indicating that barley ASL/LBD genes may have a similar function in plant development.
In plants, different aspects of root development are affected by ASL/LBD genes, including lateral and shoot-borne root development in Arabidopsis, rice, and maize (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007; Taramino et al., 2007). AtARF7 and AtARF19 regulate lateral root formation as transcriptional activators of early auxin response genes in Arabidopsis thaliana. Target-gene analysis revealed that AtARF7 and AtARF19 directly regulate the auxin-mediated transcriptions of AtASL18/LBD16 and AtASL16/LBD29 in roots, respectively. Overexpression of AtASL18/LBD16 and AtASL16/LBD29 induces lateral root formation in the absence of AtARF7 and AtARF19. In addition, AtASL20/LBD18 in conjunction with AtASL18/LBD16 functions in the initiation and emergence of lateral roots as a downstream regulator of AtARF7 and AtARF19. These data suggest that ASL/LBD genes mediate lateral root formation in Arabidopsis by different molecular pathways. In barley, the closest homologous genes of AtASL18/LBD16, AtASL16/LBD29, and AtASL20/LBD18 were HvASL16/LBD23, HvASL13/LBD18, and HvASL17/LBD24, respectively. Expression profile analysis revealed that HvASL17/LBD24 was relatively highly expressed in 4-d embryos after germination, and in roots and shoots from seedlings (10-cm shoot stage). This suggests that expression analysis may explain the conserved function in this gene family.
Besides being developmental regulators, a reverse genetic characterization of group IIa genes showed that AtASL39/LBD37, AtASL40/LBD38, and AtASL41/LBD39 mediate the repressive effect of N/NO −3 on anthocyanin biosynthesis and further affect N-responsive genes and N metabolism (Rubin et al., 2009). In the present study, HvASL21/LBD5 and HvASL23/LBD14 were expressed in all selected tissues, and highly expressed in developing tillers at the six-leaf third internode stage. The potential roles of the HvASL21/LBD5 and HvASL23/LBD14 should be further investigated.
Compliance with ethics guidelines
Bao-jian GUO, Jun WANG, Shen LIN, Zheng TIAN, Kai ZHOU, Hai-ye LUAN, Chao LYU, Xin-zhong ZHANG, and Ru-gen XU declare that they have no concfict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
List of electronic supplementary materials
Table S1 Amino acid sequence of the ASL/LBD genes in barley
Fig. S1 Promoter analysis of HvASL12/LBD12 and HvASL14/LBD19 genes
References
Artimo, P., Jonnalagedda, M., Arnold, K., et al., 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res., 40(W1):W597–W603. http://dx.doi.org/10.1093/nar/gks400
Bailey, T.L., Elkan, C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman, R., Brutlag, D., Karp, P., et al. (Eds.), Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. American Association for Artificial Intelligence, Stanford, CA, USA, p.28–36.
Borghi, L., Bureau, M., Simon, R., 2007. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell, 19(6): 1795–1808. http://dx.doi.org/10.1105/tpc.106.047159
Bortiri, E., Chuck, G., Vollbrecht, E., et al., 2006. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell, 18(3):574–585. http://dx.doi.org/10.1105/tpc.105.039032
Byrne, M.E., Barley, R., Curtis, M., et al., 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature, 408(6815):967–971. http://dx.doi.org/10.1038/35050091
Byrne, M.E., Simorowski, J., Martienssen, R.A., 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development, 129(8):1957–1965.
Evans, M.M.S., 2007. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell, 19(1):46–62. http://dx.doi.org/10.1105/tpc.106.047506
Guo, M.J., Thomas, J., Collins, G., et al., 2008. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell, 20(1):48–58. http://dx.doi.org/10.1105/tpc.107.056127
Hetz, W., Hochholdinger, F., Schwall, M., et al., 1996. Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J., 10(5):845–857. http://dx.doi.org/10.1046/j.1365-313X.1996.10050845.x
Hu, B., Jin, J.P., Guo, A.Y., et al., 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics, 31(8):1296–1297. http://dx.doi.org/10.1093/bioinformatics/btu817
Husbands, A., Bell, E.M., Shuai, B., et al., 2007. LATERAL ORGAN BOUNDARIES defines a new family of DNAbinding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res., 35(19):6663–6671. http://dx.doi.org/10.1093/nar/gkm775
International Barley Genome Sequencing Consortium, Mayer, K.F., Waugh, R., et al., 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature, 491(7426):711–716. http://dx.doi.org/10.1038/nature11543
Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., et al., 2005. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell, 17(5):1387–1396. http://dx.doi.org/10.1105/tpc.105.030981
Iwakawa, H., Ueno, Y., Semiarti, E., et al., 2002. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol., 43(5):467–478. http://dx.doi.org/10.1093/pcp/pcf077
Iwakawa, H., Iwasaki, M., Kojima, S., et al., 2007. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J., 51(2):173–184. http://dx.doi.org/10.1111/j.1365-313X.2007.03132.x
Iwasaki, M., Takahashi, H., Iwakawa, H., et al., 2013. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development, 140(9):1958–1969. http://dx.doi.org/10.1242/dev.085365
Keta, S., Iwakawa, H., Ikezaki, M., et al., 2012. Roles of the ASYMMETRIC LEAVES2 gene in floral organ development in Arabidopsis thaliana. Plant Biotechnol., 29(1):1–8. http://dx.doi.org/10.5511/plantbiotechnology.11.1101a
Koppolu, R., Anwar, N., Sakuma, S., et al., 2013. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. PNAS, 110(32):13198–13203. http://dx.doi.org/10.1073/pnas.1221950110
Lee, H.W., Kim, N.Y., Lee, D.J., et al., 2009. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol., 151(3):1377–1389. http://dx.doi.org/10.1104/pp.109.143685
Lescot, M., Déhais, P., Thijs, G., et al., 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res., 30(1):325–327. http://dx.doi.org/10.1093/nar/30.1.325
Letunic, I., Doerks, T., Bork, P., 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res., 40(D1):D302–D305. http://dx.doi.org/10.1093/nar/gkr931
Liu, R.H., Meng, J.L., 2003. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas (Beijing), 25(3):317–321.
Liu, H.J., Wang, S.F., Yu, X.B., et al., 2005. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J., 43(1):47–56. http://dx.doi.org/10.1111/j.1365-313X.2005.02434.x
Long, J.A., Moan, E.I., Medford, J.I., 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature, 379(6560):66–69. http://dx.doi.org/10.1038/379066a0
Majer, C., Hochholdinger, F., 2011. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci., 16(1):47–52. http://dx.doi.org/10.1016/j.tplants.2010.09.009
Matsumura, Y., Iwakawa, H., Machida, Y., 2009. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J., 58(3):525–537. http://dx.doi.org/10.1111/j.1365-313X.2009.03797.x
Okushima, Y., Fukaki, H., Onoda, M., et al., 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell, 19(1): 118–130. http://dx.doi.org/10.1105/tpc.106.047761
Ori, N., Eshed, Y., Chuck, G., et al., 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development, 127:5523–5532.
Punta, M., Coggill, P.C., Eberhardt, R.Y., et al., 2012. The Pfam protein families database. Nucleic Acids Res., 40(D1):D290–D301. http://dx.doi.org/10.1093/nar/gkr1065
Rubin, G., Tohge, T., Matsuda, F., et al., 2009. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell, 21(11):3567–3584. http://dx.doi.org/10.1105/tpc.109.067041
Semiarti, E., Ueno, Y., Tsukaya, H., et al., 2001. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development, 128(10):1771–1783.
Shuai, B., Reynaga-Pena, C.G., Springer, P.S., 2002. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol., 129(2):747–761. http://dx.doi.org/10.1104/pp.010926
Tamura, K., Stecher, G., Peterson, D., et al., 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol., 30(12):2725–2729. http://dx.doi.org/10.1093/molbev/mst197
Taramino, G., Sauer, M., Stauffer, J.L., et al., 2007. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and postembryonic shoot-borne root initiation. Plant J., 50(4): 649–659. http://dx.doi.org/10.1111/j.1365-313X.2007.03075.x
Wang, X.F., Zhang, S.Z., Su, L., et al., 2013. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLOS ONE, 8(2):e57044. http://dx.doi.org/10.1371/journal.pone.0057044
Xu, B., Li, Z.Y., Zhu, Y., et al., 2008. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol., 146(4):566–575. http://dx.doi.org/10.1104/pp.104.900253
Xu, C., Luo, F., Hochholdinger, F., 2016. LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci., 21(2):159–167. http://dx.doi.org/10.1016/j.tplants.2015.10.010
Yang, Y., Yu, X.B., Wu, P., 2006. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol., 39(1):248–262. http://dx.doi.org/10.1016/j.ympev.2005.09.016
Zhang, Y.M., Zhang, S.Z., Zheng, C.C., 2014. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet., 93(1):79–91. http://dx.doi.org/10.1007/s12041-014-0342-7
Author information
Authors and Affiliations
Corresponding author
Additional information
The two authors contributed equally to this work
Project supported by the National Natural Science Foundation of China (Nos. 31401370 and 31571648), the National Barley and Highland Barley Industrial Technology Specially Constructive Foundation of China (No. CARS-05), the Jiangsu Agriculture Science and Technology Innovation Fund (No. CX(12)5084), Jiangsu Training Programs of Innovation and Entrepreneurship for Undergraduates (No. 201611117090X), and the Priority Academic Program Development of Jiangsu Higher Education Institutions, China
ORCID: Bao-jian GUO, http://orcid.org/0000-0003-4147-7414
Electronic supplementary material
11585_2016_192_MOESM1_ESM.pdf
A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.), approximately 195 KB.
11585_2016_192_MOESM2_ESM.pdf
A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.), approximately 138 KB.
Rights and permissions
About this article
Cite this article
Guo, Bj., Wang, J., Lin, S. et al. A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang Univ. Sci. B 17, 763–774 (2016). https://doi.org/10.1631/jzus.B1500277
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1500277