Abstract
A Gram-negative, aerobic, non-motile, rod-shaped bacterial strain, designated 25-1T, was isolated from the air inside giant panda enclosures at the Chengdu Research Base of Giant Panda Breeding, China. Strain 25-1T grew optimally at pH 7.0–8.0, at 28–30 °C and in the presence of NaCl concentrations from 0.0% to 0.5 %. 16S rRNA gene sequence analysis indicated that strain 25-1T belongs to the genus Chryseobacterium within the family Flavobacteriaceae and is related most closely to C. carnis G81T (96.4% similarity), C. lathyri RBA2-6T (95.8% similarity), and C. zeae JM1085T (95.8% similarity). Its genomic DNA G+C molar composition was 36.2%. The major cellular fatty acids were iso-C15:0 (44.0%), iso-C17:0 3OH (19.8%) and C16:1 ω7c/16:1 ω6c (12.7%). The only isoprenoid quinone was menaquinone 6 (MK-6). The major polar lipids were phosphatidylethanolamine, two unidentified amino lipids and two unidentified lipids. The DNA-DNA relatedness between strain 25-1T and C. lathyri RBA2-6T was 38%. Phenotypic, genotypic, and phylogenetic characteristics indicated that strain 25-1T is a novel member of the genus Chryseobacterium, for which the name C. chengduensis sp. nov. is proposed. The type strain is 25-1T (CCTCC AB2015133T=DSM 100396T).
摘要
目的
鉴定菌株25-1T 是否是金黄杆菌属的一个新种。
创新点
首次从空气中分离到金黄杆菌属的新种。
方法
革兰氏染色镜检;磷钨酸染色然后透射电镜观察菌株25-1T 形态结构;全自动生理生化鉴定系统(Phoenix™-100)与传统生理生化反应管相结合;H890 气象色谱仪进行脂肪酸组分分析;薄板双相层析进行极性脂组分分析;反相高压液相色谱分析法进行呼吸琨组分分析;熔解温度法检测G+C 摩尔含量;16S rRNA 序列测定及系统发生分析。
结论
根据传统特征分类研究结果(形态特征、培养特性和生理生化特征)、化学分类研究结果(脂肪酸组分、极性脂组分和呼吸琨组分)和遗传特征分类研究结果(G+C 摩尔含量、DNA 同源性测定和16S rRNA 序列测定及系统发生分析)得出菌株25-1T 为金黄杆菌属的一个新种。
Similar content being viewed by others
1 Introduction
In this study, we investigated the cultivable bacterial community in the air of giant panda enclosures at the Chengdu Research Base of Giant Panda Breeding in Sichuan Province, located in southwestern China. Based on differences in colony morphology, 28 pure cultures were selected for 16S rRNA gene sequencing and phylogenetic analysis. Species included members of the genera Micrococcus, Chryseobacterium, Leuconostoc, Staphylococcus, Pseudomonas, Kocuria, Bacillus, Exiguobacterium, Acinetobacter, Escherichia, Rothia, and Dietzia. The strain designated 25-1T was characterized using a polyphasic taxonomy approach, including evaluation of its morphological, biochemical, and phylogenetic characteristics. Unfortunately, C. carnis G81T and C. zeae JM1085T had not been released from culture collections at the time of these investigations and so were not included as reference strains. Therefore, all tests were performed on the new isolate and on C. lathyri RBA2-6T, which was acquired from the National Institute of Technology and Evaluation (NITE) Biological Resource Center (NBRC). The data obtained revealed that strain 25-1T should be assigned to the genus Chryseobacterium as the type strain of a novel species.
2 Materials and methods
2.1 Culture conditions and phenotypic characteristics
Strain 25-1T was isolated from the cultivable bacterial community in the air of a giant panda enclosure by exposing a petri dish containing tryptic soy agar (TSA, Difco, Leeuwarden, the Netherlands) medium for 15 min. For further analysis, strain 25-1T was cultivated on Luria-Bertani (LB) agar (Difco) at 30 °C. The presence of flexirubin type pigments was investigated using a 20% (0.2 g/ml) KOH solution according to the study of Bernardet et al. (2002). Gram staining was determined using the non-staining method described by Buck (1982). Cellular morphology, motility, and other physiological characteristics were evaluated as previously described (Wen et al., 2016). Cellular morphology was observed by light microscopy (Olympus; magnification 61 000×) and cell size was determined by transmission electron microscopy (H-600-A2; Hitachi, Tokyo, Japan) using cells from an exponentially growing culture. Motility tests were performed using LB broth with 0.3% (3 g/L) agar. Growth temperatures (4, 10, 15, 20, 25, 28, 30, 37, 40, 45, and 50 °C) and pH (2.0–10.0, at intervals of 1.0 pH unit) were monitored during 7 d of incubation in LB broth as described by Xu and Wu (2005). NaCl tolerance was tested in LN medium (LB without NaCl) supplemented with 0%, 0.5%, 1.0%–5.0% (at intervals of 1%) (1%=0.01 g/ml) NaCl during 7 d of incubation. Anaerobic growth was investigated by incubation in an anaerobic chamber (Mitsubishi Gas Chemical, Tokyo, Japan) at 30 °C for 7 d on LB agar.
2.2 Biochemical characteristics and microbial sensitivity test
A number of key characteristics were tested using conventional procedures, as described by Smibert and Krieg (1994) and Skerman (1967), i.e., the production of catalase, oxidase, hydrogen sulphide and indole, and hydrolysis of Tween 80, starch, and gelatin. Some strain 25-1T and C. lathyri RBA2-6T biochemical reactions were detected using a bacterial biochemical trace kit (Hangzhou Microbial Reagent Co., Ltd., Hangzhou, China), which included the following substances: β-galactosidase, arginine decarboxylase, ornithine decarboxylase, nitrate reduction, mannose, adipic acid, arabinose, trehalose, cellobiose, lactose, salicin, and acetamide. The additional biochemical and physiological properties of strain 25-1T and C. lathyri RBA2-6T were determined using the BD Phoenix™-100 automated microbiology system (Becton Dickinson, New Jersey, USA), according to the manufacturer’s instructions. The biological principles of the Analytic Products INC (API) and Phoenix systems are similar (Wen et al., 2016), but the Phoenix system is automated and can handle a higher number of tests. Each negative identification (NID) panel contains two fluorescent positive control wells and 45 substrates (O’Hara, 2006). The sensitivity of strain 25-1T to various antibiotics was determined as previously described (Wen et al., 2016), by spreading bacterial suspensions on LB agar plates and applying filter paper discs containing the following antibiotics (ug per disc): vancomycin (30), sulfamethoxazole (100), tetracycline (30), lincomycin (10), spectinomycin (100), kanamycin (30), furazolidone (100), streptomycin (10), erythromycin (15), trimesulf (25), ciprofloxacin (5), rifampicin (5), ampicillin (10), florfenicol (30), penicillin (10), gentamicin (10), amikacin (30), cephalothin (30), ceftazidime (30), doxycycline (30), cefotaxime (30), and enrofloxacin (5) (all obtained from Hangzhou Microbial Reagents). The strain was incubated in the presence of the antibiotics at 30 °C for 24 h.
2.3 16S rRNA sequencing and phylogenetic analysis
The 16S rRNA gene of strain 25-1T was amplified by polymerase chain reaction (PCR) using two universal primers as described previously (Greisen et al., 1994), and the amplification products were sequenced by Invitrogen Biotechnology (Shanghai, China). The sequence was compared with sequences available in the EzTaxon Server (http://eztaxon-e.ezbiocloud.net; Kim O.S. et al., 2012) and GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of strain 25-1T and the type strains of published Chryseobacterium species were aligned using the Clustal_X program (Thompson et al., 1997). Phylogenetic analysis was performed using the MEGA 6.0 offline software (Tamura et al., 2013) and PHYML online web server (Guindon et al., 2005). Phylogenetic trees were constructed using the neighbor-joining, maximum-likelihood (Felsenstein, 1981), and maximum-parsimony (Kluge and Farris, 1969) methods in the MEGA 6.0 program, with bootstrap analysis based on 1000 replicates (Felsenstein, 1985).
2.4 Chemotaxonomic and genomic analyses
The DNA G+C composition of strain 25-1T was determined using the thermal denaturation method (Mandel and Marmur, 1968) with Escherichia coli K-12 as a control. Genomic DNA was extracted and purified using conventional procedures (Sambrook and Russell, 2001). Polar lipids of strain 25-1T were identified by two-dimensional thin-layer chromatography (TLC) according to the protocols of Tindall (1990). The respiratory quinones of strain 25-1T were extracted and analyzed by high performance liquid chromatography (HPLC) as described by Xie and Yokota (2003), using C. takakiae CGMCC1.13488T as a reference strain. The whole cell fatty acids of strains 25-1T and C. lathyri RBA2-6T grown on LB agar at 30 °C for 48 h were analyzed using the Sherlock microbial identification system (MIDI) and identified using the MIDI software package Version 6.0 based on the TSBA6 6.00 database. The four above-mentioned analyses were performed at the China General Microbiological Culture Collection Center (CGMCC). Genomic DNA-DNA hybridizations were carried out between strain 25-1T and C. lathyri RBA2-6T using the fluorometric method (Ezaki et al., 1989), at the Guangdong Microbiology Culture Center (GIMCC).
3 Results and discussion
3.1 Morphological and physiological characteristics
Cells of strain 25-1T were observed to be nonmotile, strictly aerobic, Gram-stain-negative, and rod-shaped (0.44–0.48 µm wide and 0.88–0.92 µm long; Fig. S1). Colonies on LB agar were nontransparent, yellow-pigmented, circular, and smooth with regular edges after 3 d of incubation at 30 °C. The non-fluorescent and non-diffusible yellow pigments belong to the flexirubin type. Cells grow in LN medium with 0%–2% NaCl with an optimum concentration of 0%–0.5%. Good growth occurs at 28–30 °C and no growth occurs below 10 °C or above 40 °C. Growth occurs at pH 6.0–9.0 and optimally at pH 7.0–8.0. The characteristics of strain 25-1T and C. lathyri RBA2-6T are shown in Tables 1 and 2. Strain 25-1 was able to use acetate, adonitol, and d-mannitol, but unable to use ornithine, sorbitol, or sucrose. In contrast to C. lathyri RBA2-6T, strain 25-1T was negative for l-glutamic acid-AMC, γ-glutamyl-NA, l-proline-NA, bis-pNP-phosphate (pNP: p-nitropheno), pNP-β-d-glucoside, and esculin hydrolysis.
3.2 Phylogenetic analysis
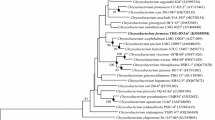
The 16S rRNA gene sequence (1401 bp) of strain 25-1T was obtained (GenBank accession number KP966546). Strain 25-1T showed less than 96.4% sequence similarity to the type strains of all recognized species in the genus Chryseobacterium. The highest 16S rRNA sequence similarity was found with C. carnis NCTC 13525T (96.4%) (Holmes et al., 2013), followed by C. lathyri RBA2-6T (95.8%), C. zeae JM1085T (95.8%) (Kämpfer et al., 2014b), C. shigense GUM kajiT (95.8%) (Shimomura et al., 2005), C. gwangjuense THG A-18T (95.7%) (Park et al., 2013), and C. carnipullorum 9-R23581T (95.7%) (Charimba et al., 2013), as determined using the EzTaxon server 2.1. Strain 25-1T was included in a cluster containing the type strains of C. carnis and C. chaponense, forming a distinct phylogenetic lineage within the genus Chryseobacterium in the neighbor-joining phylogenetic tree (Fig. 1). The phylogenetic position was also supported by the maximum-parsimony and maximum-likelihood trees.
Phylogenetic tree showing the relationship between strain 25-1 T and the type strains of a selection of recognized Chryseobacterium species constructed by the neighbor-joining method based on their 16S rRNA gene sequences
GenBank accession numbers are shown in parentheses. Numbers indicate percentages of occurrence of branch points in 1000 bootstrapped trees. The bar is 0.5% in the legend and 0.005 on the tree
3.3 Chemotaxonomic and genomic characteristics
The cellular fatty acids of strain 25-1T and C. lathyri RBA2-6T analyzed under the same conditions are shown in Table 3. The predominant fatty acids of strain 25-1 (≥5%) were iso-C15:0 (44.0%), iso-C17:0 3OH (19.8%), summed feature 3 (C16:1 ω7c and/or C16:1 ω6c, 12.7%) and summed feature 9 (10-methyl C16:0 and/or iso-C17:1 ω9c, 7.8%). Minor amounts of anteiso-C15:0 (1.4%), iso-C16:0 (1.8%), iso-C15:0 3OH (3.9%), iso-C16.0 3OH (2.0%), and d16.0 3OH (1.2%) were also detected. The presences of major fatty acids, namely iso-C15:0, iso-C17:0 3OH and iso-C17:1 ω9c, are in accordance with the placement of strain 25-1T in the genus Chryseobacterium (Li and Zhu, 2012; Kämpfer et al., 2015b). However, strain 25-1T could be readily distinguished from C. lathyri by the presence of a high amount of iso-C17:0 3OH and a significantly lower amount of summed feature 9. The discrepancies noted in the fatty acid composition of C. lathyri RBA2-6T as determined in this study compared to the data reported in the original description (Cho et al., 2010) may be due to differences in the fatty acid extraction methods, types of gas chromatography, or the culture media used. The isoprenoid quinone of strain 25-1T was MK-6, which is characteristic of all members of the family Flavobacteriaceae (Kämpfer et al., 2009). The polar lipids of strain 25-1T were phosphatidylethanolamine (PE), two unidentified amino lipids and two unidentified lipids (Fig. 2), which is in line with other recognized species of the genus Chryseobacterium (Kämpfer et al., 2015a). The DNA G+C molar content of strain 25-1T was 36.2%. This value is within the range reported for Chryseobacterium species (Bernardet et al., 2010; Montero-Calasanz et al., 2014). The mean DNA-DNA relatedness between strain 25-1T and C. lathyri RBA2-6T was 38%. This is clearly far below the 70% threshold value that is generally used for prokaryotic species delineation (Stackebrandt et al., 2002).
Total polar lipid analysis of strain Chryseobacterium chengduensis 25-1 T carried out by two-dimensional thinlayer chromatography (TLC)
Ascending solvent system: 1st dimension: chloroform/methanol/water (65:25:4, v/v); 2nd dimension: chloroform/methanol/acetic acid/water (80:12:15:4, v/v). Molybdatophosphoric acid was applied for the detection of polar lipids. PE, phosphatidylethanolamine; AL1 and AL2, unidentified amino lipids; UL1 and UL2, unidentified lipids
4 Conclusions
The physiological, chemotaxonomic, and phylogenetic analyses conducted in this study show that strain 25-1T exhibits the main properties of the genus Chryseobacterium but can be differentiated from the closely related type strain, C. lathyri RBA2-6T. The distinctiveness of strain 25-1 is sufficient to categorize the isolate as a member of a species that is distinguished from the published Chryseobacterium species. The low 16S rRNA gene sequence similarity with all other described Chryseobacterium species supports the description of strain 25-1 as a member of a new Chryseobacterium species for which we propose the name C. chengduensis sp. nov. (cheng.du.en’sis. N.L. fem. adj. chengduensis pertaining to Chengdu in Sichuan Province, China, where the type strain was isolated). Cells of strain 25-1T are nonmotile, strictly aerobic, Gram-stain-negative, and rod-shaped (0.44–0.48 µm wide and 0.88–0.92 µm long). Colonies on LB agar are non-transparent, yellow-pigmented, circular, and smooth with regular edges after 3 d of incubation at 30 °C. The non-fluorescent and non-diffusible yellow pigments belong to the flexirubin type. Cells grow in LN medium with 0%–2% NaCl with an optimum concentration of 0%–0.5%. Optimal growth occurs at 28–30 °C and no growth occurs below 10 °C or above 40 °C. Growth occurs at pH 6.0–9.0 and optimally at pH 7.0–8.0. Catalase and oxidase are produced, but not indole or H2S. Starch and Tween 80 are degraded but not gelatin. Nitrate is not reduced to nitrite. Acid is not produced from salicin, acetamide, mannose, β-galactosidase, trehalose, cellobiose, arabinose, and lactose. It is negative for adipic acid and arginine decarboxylase, and positive for the production of ornithine decarboxylase. In the Phoenix system, it is positive for arginine-arginine-AMC, glycine-proline-AMC, glycine-AMC, glutaryl-glycine-arginine-AMC, l-arginine-AMC, l-leucine-AMC, l-phenylalanine-AMC, l-pyroglutamic acid-AMC, l-tryptophan-AMC, lysine-alanine-AMC, acetate, adonitol, citrate, colistin, d-mannitol, α-ketoglutaric acid, malonate, tiglic acid, polymyxin B, and 4MU-#-acetyl-β-d-glucosaminide. It is sensitive to vancomycin, tetracycline, rifampicin, ciprofloxacin, florfenicol, doxycycline, amikacin, enrofloxacin, cefotaxime, ceftazidime, furazolidone, and trimesulf, but resistant to kanamycin, spectinomycin, erythromycin, cephalothin, sulfamethoxazole, lincomycin, streptomycin, ampicillin, penicillin, and gentamicin. The major cellular fatty acids are iso-C15:0, iso-C17:0 3OH, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c) and summed feature 9 (10-methyl C16:0 and/or iso-C17:1 ω9c). The only isoprenoid quinone detected was MK-6. The DNA G+C molar content was 36.2%. The major polar lipids are phosphatidylethanolamine, two unidentified amino lipids and two unidentified lipids.
The type strain, 25-1T (CCTCC AB2015133T=DSM 1003961), was isolated from the air inside giant panda enclosures at the Chengdu Research Base of Giant Panda Breeding in Sichuan Province, China.
Compliance with ethics guidelines
Cai-fang WEN, Li-xin XI, Shan ZHAO, Zhong-xiang HAO, Lu LUO, Hong LIAO, Zhen-rong CHEN, Rong SHE, Guo-quan HAN, San-jie CAO, Rui WU, Qi-gui YAN, and Rong HOU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
List of electronic supplementary materials
Fig. S1 Transmission electron micrograph of a negatively stained cell of strain 25-1T, showing the absence of flagella
References
Bernardet, J.F., Nakagawa, Y., Holmes, B., 2002. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol., 52(3):1049–1070. http://dx.doi.org/10.1099/00207713-52-3-1049
Bernardet, J.F., Hugo, C.J., Bruun, B., 2010. Chryseobacterium Vandamme Bernardet, Segers, Kersters and Holmes 1994, 829VP. In: Krieg, N.R., Staley, J.T., Brown, D.R. (Eds.), Bergey’s Manual® of Systematic Bacteriology, 2nd Ed., Springer New York, p.180–196. http://dx.doi.org/10.1007/978-0-387-68572-4
Buck, J.D., 1982. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol., 44(4):992–993.
Charimba, G., Jooste, P., Albertyn, J., et al., 2013. Chryseobacterium carnipullorum sp. nov., isolated from raw chicken. Int. J. Syst. Evol. Microbiol., 63(Pt 9):3243–3249. http://dx.doi.org/10.1099/ijs.0.049445-0
Chaudhari, P.N., Wani, K.S., Chaudhari, B.L., et al., 2009. Characteristics of sulfobacin A from a soil isolate Chryseobacterium gleum. Appl. Biochem. Biotechnol., 158(1):231–241. http://dx.doi.org/10.1007/s12010-008-8417-7
Cho, S.H., Lee, K.S., Shin, D.S., et al., 2010. Four new species of Chryseobacterium from the rhizosphere of coastal sand dune plants, Chryseobacterium elymi sp. nov., Chryseobacterium hagamense sp. nov., Chryseobacterium lathyri sp. nov. and Chryseobacterium rhizosphaerae sp. nov. Syst. Appl. Microbiol., 33(3):122–127. http://dx.doi.org/10.1016/j.syapm.2009.12.004
Ezaki, T., Hashimoto, Y., Yabuuchi, E., 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol., 39(3):224–229. http://dx.doi.org/10.1099/00207713-39-3-224
Greisen, K., Loeffelholz, M., Purohit, A., et al., 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol., 32(2):335–351.
Felsenstein, J., 1981. Evolutionary trees from DNA sequences:a maximum likelihood approach. J. Mol. Evol., 17(6):368–376. http://dx.doi.org/10.1007/BF01734359
Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39(4):783–791. http://dx.doi.org/10.2307/2408678
Guindon, S., Lethiec, F., Duroux, P., et al., 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res., 33(Suppl. 2):W557–W559. http://dx.doi.org/10.1093/nar/gki352
Holmes, B., Steigerwalt, A.G., Nicholson, A.C., 2013. A DNA-DNA hybridization study of strains of Chryseobacterium, Elizabethkingia, Empedobacter and of other usually indole-producing non-fermenters of CDC groups IIc, IIe, IIh and IIi, all from mostly human clinical sources and proposals of Chryseobacterium bernardetii sp. nov., Chryseobacterium carnis sp. nov., Chryseobacterium lactis sp. nov., Chryseobacterium nakagawai sp. nov., and Chryseobacterium taklimakanense comb. nov. Int. J. Syst. Evol. Microbiol., 63(Pt 12):4639–4662. http://dx.doi.org/10.1099/ijs.0.054353-0
Ilardi, P., Fernández, J., Avendaño-Herrera, R., 2009. Chryseobacterium piscicola sp. nov., isolated from diseased salmonid fish. Int. J. Syst. Evol. Microbiol., 59(12):3001–3005. http://dx.doi.org/10.1099/ijs.0.007021-0
Kämpfer, P., Vaneechoutte, M., Lodders, N., et al., 2009. Description of Chryseobacterium anthropi sp. nov. to accommodate clinical isolates biochemically similar to Kaistella koreensis and Chryseobacterium haifense, proposal to reclassify Kaistella koreensis as Chryseobacterium koreense comb. nov. and emended description of the genus Chryseobacterium. Int. J. Syst. Evol. Microbiol., 59(10):2421–2428. http://dx.doi.org/10.1099/ijs.0.008250-0
Kämpfer, P., Poppel, M.T., Wilharm, G., 2014a. Chryseobacterium gallinarum sp. nov., isolated from a chicken, and Chryseobacterium contaminans sp. nov., isolated as a contaminant from a rhizosphere sample. Int. J. Syst. Evol. Microbiol., 64(Pt 4):1419–1427. http://dx.doi.org/10.1099/ijs.0.058933-0
Kämpfer, P., McInroy, J.A., Glaeser, S.P., 2014b. Chryseobacterium zeae sp. nov., Chryseobacterium arachidis sp. nov., and Chryseobacterium geocarposphaerae sp. nov. isolated from the rhizosphere environment. Antonie van Leeuwenhoek, 105(3):491–500. http://dx.doi.org/10.1007/s10482-013-0101-4
Kämpfer, P., Busse, H.J., McInroy, J.A., et al., 2015a. Chryseobacterium arachidiradicis sp. nov. isolated from the geocarposphere (soil around the peanut) of very immature peanuts (Arachis hypogaea). Int. J. Syst. Evol. Microbiol., 65(7):2179–2186. http://dx.doi.org/10.1099/ijs.0.000237
Kämpfer, P., Trček, J., Skok, B., et al., 2015b. Chryseobacterium limigenitum sp. nov., isolated from dehydrated sludge. Antonie van Leeuwenhoek, 107(6):1633–1638. http://dx.doi.org/10.1007/s10482-015-0434-2
Kim, H.S., Sang, M.K., Jung, H.W., et al., 2012. Identification and characterization of Chryseobacterium wanjuense strain KJ9C8 as a biocontrol agent of Phytophthora blight of pepper. Crop Prot., 32:129–137. http://dx.doi.org/10.1016/j.cropro.2011.10.018
Kim, O.S., Cho, Y.J., Lee, K., et al., 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol., 62(Pt 3):716–721. http://dx.doi.org/10.1099/ijs.0.038075-0
Kluge, A.G., Farris, J.S., 1969. Quantitative phyletics and the evolution of anurans. Syst. Zool., 18(1):1–32. http://dx.doi.org/10.2307/2412407
Li, Z., Zhu, H., 2012. Chryseobacterium vietnamense sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol., 62(Pt 4):827–831. http://dx.doi.org/10.1099/ijs.0.027201-0
Lo, H.H., Chang, S.M., 2014. Identification, characterization, and biofilm formation of clinical Chryseobacterium gleum isolates. Diagn. Micr. Infec. Dis., 79(3):298–302. http://dx.doi.org/10.1016/j.diagmicrobio.2014.01.027
Loch, T.P., Faisal, M., 2014. Chryseobacterium aahli sp. nov., isolated from lake trout (Salvelinus namaycush) and brown trout (Salmo trutta), and emended descriptions of Chryseobacterium ginsenosidimutans and Chryseobacterium gregarium. Int. J. Syst. Evol. Microbiol., 64(Pt 5):1573–1579. http://dx.doi.org/10.1099/ijs.0.052373-0
Mandel, M., Marmur, J., 1968. Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol., 12(Pt B):195-206. http://dx.doi.org/10.1016/0076-6879(67)12133-2
Monteen, M.R., Ponnapula, S., Wood, G.C., et al., 2013. Treatment of Chryseobacterium indologenes ventilatorassociated pneumonia in a critically Ill trauma patient. Ann. Pharmacother., 47(12):1736–1739. http://dx.doi.org/10.1177/1060028013508745
Montero-Calasanz, M., Göker, M., Rohde, M., et al., 2014. Chryseobacterium oleae sp. nov., an efficient plant growth promoting bacterium in the rooting induction of olive tree (Olea europaea L.) cuttings and emended descriptions of the genus Chryseobacterium, C. daecheongense, C. gambrini, C. gleum, C. joostei, C. jejuense, C. luteum, C. shigense, C. taiwanense, C. ureilyticum and C. vrystaatense. Syst. Appl. Microbiol., 37(5):342–350. http://dx.doi.org/10.1016/j.syapm.2014.04.004
Nemli, S.A., Demirdal, T., Ural, S., 2015. A case of healthcare associated pneumonia caused by Chryseobacterium indologenes in an immunocompetent patient. Case Report. Infect. Dis., 2015:483923. http://dx.doi.org/10.1155/2015/483923
O’Hara, C.M., 2006. Evaluation of the Phoenix 100 ID/AST system and NID panel for identification of Enterobacteriaceae, Vibrionaceae, and commonly isolated nonenteric Gram-negative bacilli. J. Clin. Microbial., 44(3):928–933. http://dx.doi.org/10.1128/JCM.44.3.928-933.2006
Park, Y.J., Son, H.M., Lee, E.H., et al., 2013. Chryseobacterium gwangjuense sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol., 63(Pt 12):4580–4585. http://dx.doi.org/10.1099/ijs.0.052118-0
Sambrook, J., Russell, D.W., 2001. Molecular Cloning: A Laboratory Manual, 3rd Ed. Cold Spring Harbor, NY.
Sharma, P., Gupta, S.K., Diene, S.M., et al., 2015. Wholegenome sequence of Chryseobacterium oranimense, a colistin resistant bacterium isolated from a cystic fibrosis patient in France. Antimicrob. Agents Chemother., 59(3):1696–1706. http://dx.doi.org/10.1128/AAC.02417-14
Shimomura, K., Kaji, S., Hiraishi, A., 2005. Chryseobacterium shigense sp. nov., a yellow-pigmented, aerobic bacterium isolated from a lactic acid beverage. Int. J. Syst. Evol. Microbiol., 55(5):1903–1906. http://dx.doi.org/10.1099/ijs.0.63690-0
Skerman, V.B.D., 1967. A Guide to the Identification of the Genera of Bacteria, 2nd Ed. Williams & Wilkins Co., Baltimore.
Smibert, R.M., Krieg, N.R., 1994. Phenotypic characterization. In: Gerhardt, P., Murray, R.G.E., Wood, W.A., et al. (Eds.), Methods for General and Molecular Bacteriology, American Society for Microbiology. Washington, DC, p.607–653.
Stackebrandt, E., Frederiksen, W., Garrity, G.M., et al., 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol., 52(3):1043–1047. http://dx.doi.org/10.1099/00207713-52-3-1043
Tamura, K., Stecher, G., Peterson, D., et al., 2013. MEGA6:Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol., 30(12):2725–2729. http://dx.doi.org/10.1093/molbev/mst197
Thompson, J.D., Gibson, T.J., Plewniak, F., et al., 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25(24):4876–4882. http://dx.doi.org/10.1093/nar/25.24.4876
Tindall, B.J., 1990. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett., 66(1–3):199–202. http://dx.doi.org/10.1111/j.1574-6968.1990.tb03996.x
Vandamme, P., Bernardet, J.F., Segers, P., et al., 1994. New perspectives in the classification of the flavobacteria:description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int. J. Syst. Bacteriol., 44(4):827–831. http://dx.doi.org/10.1099/00207713-44-4-827
Wang, S.L., Liang, Y.C., Liang, T.W., 2011. Purification and characterization of a novel alkali-stable α-amylase from Chryseobacterium taeanense TKU001, and application in antioxidant and prebiotic. Process Biochem., 46(3):745–750. http://dx.doi.org/10.1016/j.procbio.2010.11.022
Wen, C., Xi, L., She, R., et al., 2016. Lysobacter chengduensis sp. nov. isolated from the air of captive Ailuropoda melanoleuca enclosures in Chengdu, China. Curr. Microbiol., 72(1):88–93. http://dx.doi.org/10.1007/s00284-015-0921-8
Xie, C.H., Yokota, A., 2003. Phylogenetic analysis of Lampropedia hyalina based on the 16S rRNA gene sequence. J. Gen. Appl. Microbiol., 49(6):345–349. http://dx.doi.org/10.2323/jgam.49.345
Xu, X.W., Wu, M., 2005. Isolation and characterization of a novel strain of Natrinema containing a bop gene. J. Zhejiang Univ.-Sci. B (Biomed. & Biotechnol.), 6(2):142–146. http://dx.doi.org/10.1631/jzus.2005.B0142
Yang, F., Liu, H.M., Li, S.P., et al., 2015. Chryseobacterium shandongense sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol., 65(6):1860–1865. http://dx.doi.org/10.1099/ijs.0.000186
Author information
Authors and Affiliations
Corresponding author
Additional information
The two authors contributed equally to this work
Project supported by the Chengdu Giant Panda Breeding Research Foundation (No. CPF2010-06) and the National Key Technology R & D Program of China (No. 2012BAC01B06)
Electronic supplementary materials: The online version of this article (http://dx.doi.org/10.1631/jzus.B1500214) contains supplementary materials, which are available to authorized users
ORCID: Qi-gui YAN, http://orcid.org/0000-0001-5721-2432
Electronic supplementary material
11585_2016_35_MOESM1_ESM.pdf
Fig. S1 Transmission electron micrograph of a negatively stained cell of strain 25-1T, showing the absence of flagella, approximately 81 KB.
Rights and permissions
About this article
Cite this article
Wen, Cf., Xi, Lx., Zhao, S. et al. Chryseobacterium chengduensis sp. nov. isolated from the air of captive giant panda enclosures in Chengdu, China. J. Zhejiang Univ. Sci. B 17, 610–618 (2016). https://doi.org/10.1631/jzus.B1500214
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1500214