Abstract
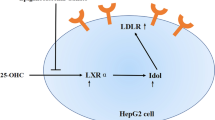
We studied the effects of alfalfa saponin extract (ASE) on low density lipoprotein receptor (Ldlr), liver X receptor a (LXRα), and farnesoid X receptor (FXR) in normal and hyperlipidemic Buffalo rat liver (BRL) cells. Normal and hyperlipidemic BRL cells were divided into eight groups: normal, or normal cells treated with 50, 100, and 150 mg/L ASE, hyperlipidemic, or hyperlipidemic cells treated with 50, 100, and 150 mg/L ASE. After treatment for 24 h, Ldlr, LXRα, and FXR mRNA expression levels were measured by quantitative real-time polymerase chain reaction (qRT-PCR). Data showed that mRNA expression of Ldlr in normal BRL cells was significantly up-regulated by ASE treatment and mRNA expressions of LXRα and FXR were significantly down-regulated both in normal and hyperlipidemic BRL cells after ASE treatment. Thus, ASE might ameliorate hepatic steatosis by regulating genes involved in cholesterol metabolism, including up-regulation of Ldlr as well as down-regulation of LXRα and FXR.
概要
目的
分析苜蓿皂苷提取物对正常和脂变Buffalo大鼠肝细胞 (BRL细胞) 中Ldlr、LXRα和FXR mRNA表达的影响, 进而初步探讨苜蓿皂苷调节脂质代谢及降低机体胆固醇含量的机制。
创新点
苜蓿皂苷具有调节脂质代谢, 降低机体胆固醇含量的作用, 但是目前对其机理研究并不多, 特别是在细胞水平上。本试验以BRL细胞为对象, 研究苜蓿皂苷对胆固醇代谢相关基因表达的影响, 从而探究苜蓿皂苷在细胞水平上对胆固醇代谢的调节作用。
方法
通过50%胎牛血清诱导BRL细胞48 h建立细胞脂肪变性模型, 运用实时荧光定量聚合酶链式反应 (qRT-PCR) 检测正常和脂变BRL细胞Ldlr、LXRα和FXR mRNA的表达, 从而得出各处理组BRL细胞内Ldlr、LXRα和FXR mRNA表达的变化。
结论
(1) 添加苜蓿皂苷后, 正常BRL细胞中Ldlr mRNA的表达量显著上调, 脂变BRL细胞Ldlr mRNA的表达有增加的趋势; (2) 苜蓿皂苷可以显著下调正常和脂变BRL细胞中LXRα和FXR mRNA的表达。因此, 在细胞水平上, 苜蓿皂苷可能通过促进Ldlr的表达, 抑制LXRα和FXR的表达, 从而调节BRL细胞胆固醇的代谢。
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Recently, as global industrialization has increased, high cholesterol levels, atherosclerosis, coronary heart disease, and other cardiovascular diseases have been increasingly prevalent threats to human health (Lopez et al., 2006). High total cholesterol (TC), triglycerides (TG), and low-density lipoprotein cholesterol (LDL-C) are thought to be independent heritable risk factors for cardiovascular diseases (Kathiresan et al., 2007). Because several plants are reported to reduce cholesterol, new plant agents with lipid-reducing activity have been under study. Specifically, alfalfa saponin extract (ASE) from alfalfa (Medicago sativa L.) contains saponins comprising a sugar moiety glycosidically linked to a hydrophobic aglycone (sapogenin), mainly pentacyclic triterpenoid (Sen et al., 1998). Because ASE has diverse biological functions for regulating lipid and cholesterol metabolism, and immune activity, it is under investigation for lipid-lowering capabilities. Alfalfa saponin (AS) and alfalfa seeds have been documented to reduce hypercholesterolemia, aortic sudanophilia, and lower cholesterol in the aortic intima-plus-media and liver (Malinow et al., 1980). ASE also prevents increased plasma cholesterol associated with a semipurified high-butter, high-cholesterol diet in monkeys (Mallnow et al., 1977). The literature suggests that AS can significantly reduce serum TC, TG, and LDL-C in animals (Al-Bar et al., 1993; Francis et al., 2001; Santoso et al., 2004) and similar outcomes in weaning piglets were reported by Wang (2007). Shi et al. (2014) concluded that the cholesterol-lowering effects of ASE were mediated by regulating gene expression involved in cholesterol metabolism. However, how ASE works at the cellular level is unclear.
Low density lipoprotein receptor (LDLR), liver X receptor α (LXRα), and farnesoid X receptor (FXR) play a vital role in cholesterol metabolism. In addition, our earlier studies in hyperlipidemic rats indicated that these three genes (Ldlr, LXRα, and FXR) were of great significance in ameliorating hyperlipidemia after ASE treatment. Thus, we sought to investigate the effects of ASE on cholesterol metabolism at a cellular level by measuring mRNA expression of Ldlr, LXRα, and FXR in normal and hyperlipidemic Buffalo rat liver (BRL) cells using quantitative real-time polymerase chain reaction (qRT-PCR).
2 Materials and methods
2.1 Materials
ASE was provided by Hebei Bao’en Biotechnology Co., Ltd. (Shijiazhuang, China). This was obtained by ethanol extraction and macroporous resin column separation and purification, and the purity was 62%. ASE was prepared as follows: powdered dried leaves and stems of alfalfa were defatted by Soxhlet extraction with petroleum ether (2×24 h) and the defatted powder was extracted with 75% ethanol (10 ml/g) for 3 h with constant stirring. After suction filtration, the extraction was repeated, and the extracts were combined and evaporated under vacuum. The dried extracts were dissolved in distilled water at a concentration of 100 g/L and then fractionated on a macroporous adsorption resin AB-8 column (Hebei Bao’en Biotechnology Co., Ltd., Shijiazhuang, China) with 50% ethanol. Ethanol-extracted saponins were obtained from 50% ethanol fractions and evaporated under vacuum. BRL cells were obtained from the Shanghai Institute of Cell Resource Center of Life Science (Shanghai, China).
2.2 Cell culture
BRL cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere with 5% CO2 for maintenance. The medium was changed every two days and culture bottles were regularly observed microscopically. BRL cells were subcultured or frozen when grown to around 80% of the bottle bottom.
2.3 Growth inhibition assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to measure the cell viability after being treated with different levels of ASE. Briefly, BRL cells (2×103 cells/well) were seeded onto a 96-well plate (Corning, NY) and cultured for 24 h at 37 °C and then treated with various concentrations of ASE (0, 30, 60, 90, 120, 150, 180, and 210 mg/L) for 24 h, six wells for each group. Afterward, MTT solution (5 mg/ml in phosphate-buffered saline (PBS)) was added (10 μl/well) and cells were incubated for another 4 h. The purple formazan crystals were dissolved in 150 μl dimethyl sulfoxide (DMSO). Then, the optical density (OD) value was detected at a wavelength of 570 nm with no cells blank hole as zero.
2.4 Hyperlipidemic cell model and grouping
About 1.5×105 subcultured BRL cells/well were seeded onto a 6-well plate, with 2 ml DMEM containing 10% FBS in every well. After cultivation for 48 h, the medium was changed and cells were randomly divided into four groups, with six replicates in each group as follows: normal control groups of 0, 0.5, 1.0, and 1.5 ml ASE solution (200 mg/L) with 2.0, 1.5, 1.0, and 0.5 ml DMEM containing 10% FBS. After treatment with different concentrations of ASE for 24 h, cells were collected for assay. Then, about 1.5×105 subcultured BRL cells/well were seeded onto 6-well plates, and grown to about 80% of the bottle bottom. Then the medium was changed into 2 ml DMEM containing 50% FBS. After cultivation for 48 h, lipid droplets or bubble deposits within the cells formed and the extent of adipose conversion was quantified by staining accumulated lipid with Oil red O and measuring extracted dye at 510 nm (Ramírez-Zacarías et al., 1992). When the hyperlipidemic BRL cell model was established, BRL cells were then placed in DMEM containing 0.1% FBS for 24 h. Next, they were randomly divided into four groups, with six replicates in each group and the normal BRL cells as the control group, and the groups were as follows: normal control group: 0 ml ASE solution (200 mg/L) with 2 ml DMEM containing 10% FBS; hyperlipidemic cells: 0, 0.5, 1.0, and 1.5 ml ASE solution (200 mg/L) with 2.0, 1.5, 1.0, and 0.5 ml DMEM containing 10% FBS. After treatment with different ASE concentrations for 24 h, cells were collected for assay.
2.5 Sample preparation and analysis
Collected BRL cells were homogenized in TRIzol reagent (Invitrogen, USA) and total RNA was isolated according to the manufacturer’s recommendations. RNA was quantified by measuring the OD of samples at 260 and 280 nm. Then samples were placed on a 1% (0.01 g/ml) agarose gel and visualized with ethidium bromide. Primers for Ldlr, LXRα, FXR (target gene) and glyceraldehydes-3-phosphate dehydrogenase (GAPDH, endogenous and stable reference gene) were designed by Primer Premier 5.0 (Canada). Primers of Ldlr gene (forward 5'-CGGCTA CCGTGAATACAGG-3', reverse 5'-GGACAAGTCG GACGAGGAG-3'), LXRα gene (forward 5'-CTCGG ATGACTCCAACCC-3', reverse 5'-CGCAGTGCCT GATGTTTC-3'), FXR gene (forward 5'-TCAGCCAA CATTCCCATC-3', reverse 5'-CCTGTGACAAAGAA GCCG-3'), and GAPDH gene (forward 5'-TCCACCC TGTTGCTGTA-3', reverse 5'-AAGAAGGTGGTG AAGCAGGC-3') for rats were synthesized by Shanghai Sangon Biological Engineering Technology & Service Co. Ltd., China.
2.6 Quantitative real-time PCR
Complementary DNA (cDNA) was synthesized with Reverse Transcriptase M-MLV (RNase H-) (TaKaRa, Dalian, China; Code No. D2639A) using the oligo dT. Real-time PCR technology was employed to determine the mRNA levels of Ldlr, LXRα, and FXR on the LightCycler instrument (Roche Diagnostics, Germany) using the SYBR Green method. Each PCR mixture (final volume of 20 μl) was composed of 10 μl of SYBR qPCR Mix (TaKaRa, Dalian, China), 0.4 μl of each gene-specific primer as mentioned above, and 1 μl cDNA in each reaction. Thermal cycling parameters were as follows: 95 °C for 120 s, followed by 40 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 30 s. Relative expression of target genes was calculated using the
method (Livak and Schmittgen, 2001).
2.7 Statistical analysis
All results were expressed as mean±standard deviation (SD). Data were evaluated by one-way analysis of variance (ANOVA; SPSS for Windows, Version 19.0, IBM, USA), and the differences between the means were assessed using Duncan’s test. Differences were considered significant at the level of P<0.05 and P<0.01 was established for very significant differences.
3 Results
3.1 Effects of ASE on cell viability
Fig. 1 showed that there were no significant effects on cell proliferation with ASE at 0, 30, 60, 90, 120, or 150 mg/L (P>0.05). BRL cell proliferation changed significantly with ASE of 180 and 210 mg/L (P<0.01).
3.2 Effects of ASE on mRNA expression of Ldlr, LXRα, and FXR in normal BRL cells
As shown in Fig. 2, mRNA expression of Ldlr in ASE groups was elevated compared with controls (P<0.01). mRNA expression of Ldlr in the ASE 150 mg/L group was greater than those treated with ASE 50 and 100 mg/L (P<0.01), which were not different from one another (P>0.05). Compared with the normal control group, mRNA expression of LXRa and FXR in ASE 100 and 150 mg/L groups decreased very significantly (P<0.01), and expression in the ASE 50 mg/L group reduced significantly (P<0.05). ASE 50, 100, and 150 mg/L groups were not different from one another (P>0.05).
3.3 Effects of ASE on mRNA expression of Ldlr, LXRα, and FXR in hyperlipidemic cells
As summarized in Fig. 3, mRNA expression of Ldlr in the hyperlipidemic cell and ASE groups decreased markedly compared with normal controls (P<0.01). There was a linear and slight rise of mRNA expression of Ldlr in hyperlipidemic cells as ASE increased (P>0.05). Compared with the normal control group, mRNA expression of LXRa in the hyperlipidemic cell group was significantly up-regulated (P<0.05), while that in the hyperlipidemic ASE 150 mg/L group was significantly down-regulated (P<0.05). Compared with the hyperlipidemic cell group, mRNA expression of LXRa in hyperlipidemic cells decreased linearly with increased ASE (P<0.01). mRNA expression of FXR in the hyperlipidemic cell group increased significantly compared with the normal control group (P<0.01). However, mRNA expression of FXR in hyperlipidemic ASE groups decreased remarkably compared with the hyperlipidemic cell group (P<0.01). There was no difference among hyperlipidemic ASE groups and the normal control group (P>0.05).
4 Discussion
4.1 Effects of ASE on cell viability
MTT assays are used to measure cytotoxicity, cell viability, and proliferation studies in cell biology (Stockert et al., 2012), so MTT assay was used to evaluate the effects of different concentrations of ASE on BRL cells. BRL cells were tolerant to ASE at 0, 30, 60, 90, 120, and 150 mg/L and were susceptible to ASE at 180 and 210 mg/L, so ASE at 50, 100, and 150 mg/L was selected for subsequent experiments.
4.2 Effects of ASE on mRNA expression of Ldlr in BRL cells
Elevated circulating LDL-C is a risk factor for cardiovascular disease (Bittner, 2003). Excess LDL-C is deposited into blood vessel walls to initiate and promote atherosclerosis and becomes a major component of the cause of atherosclerotic plaques (Pischon et al., 2005). Clearance of plasma LDL-C is primarily mediated by hepatic LDLR (Brown and Goldstein, 1983). LDLR is a cell surface glycoprotein that plays a vital role in hepatic uptake and clearance of plasma cholesterol (Brown and Goldstein, 1986). With abnormal LDLR structure or function, plasma cholesterol increases and can form plaques causing premature coronary heart disease. Thus, modulating plasma LDL-C via up-regulation of LDLR expression using drugs or plant extracts may provide therapeutic benefit.
Previous studies suggested that many plants contain compounds that can modulate plasma LDL-C via up-regulation of Ldlr expression. At low micromolar concentrations, the plant flavonoid quercetin-3-β-D-glucoside (Q3G) increased Ldlr expression and stimulated LDL uptake in human Huh7 hepatocytes (Mbikay et al., 2014). N-butanol extract (NE3) of Panax notoginseng (Burk.) F.H. Chen root significantly decreased concentrations of serum TC, TG, and LDL-C and increased Ldlr mRNA in rats (Ji and Gong, 2007). Vanillin rich fraction (VRF) extracted from vanilla pods increased Ldlr mRNA in a dose-dependent manner in HepG2 cells (Al-Naqeb et al., 2010). Berberine (BBR), a compound isolated from a Chinese herb, was observed to increase Ldlr expression and reduce serum cholesterol and LDL-C in hyperlipidemic hamsters (Kong et al., 2004), with similar effects observed in HepG2 cells (Abidi et al., 2006; Li et al., 2009).
In the current study, ASE significantly up-regulated mRNA expression of Ldlr in normal BRL cells. With increasing concentrations of ASE, mRNA expression of Ldlr increased linearly, indicating that the Ldlr had a dosage-dependent relationship with ASE. Compared with the normal control group, mRNA expression of Ldlr in the hyperlipidemic cell group was markedly down-regulated, which may be related to the formation of hepatocyte steatosis. Treated with ASE, the mRNA expression of Ldlr in hyperlipidemic BRL cells showed a linear and slight rise trend with increased ASE. A deficiency of LDLR causes LDL to accumulate as a result both of overproduction and of delayed removal (Rudling, 1992). cmice presented a significant increase in cholesterol (Evangelho et al., 2011). Increased LDLR expression or activity can reduce serum LDL cholesterol by enhancing the uptake and clearance (Reena et al., 2011). In our study, Ldlr mRNA increased both in normal and hyperlipidemic BRL cells, indicating that ASE could ameliorate hepatocyte steatosis by up-regulating Ldlr. These data agree with our previous study showing that the cholesterol-lowering effects of ASE may be attributed to its regulation on some key genes involved in cholesterol metabolism in hyperlipidemic rats, such as up-regulation of Ldlr in the liver of hyperlipidemic rats (Shi et al., 2014).
4.3 Effects of ASE on mRNA expression of LXRa in BRL cells
Cholesterosis is a disease of cholesterol metabolism characterized by the presence of excessive lipid droplets in the cytoplasm. These lipid droplets are mainly composed of cholesterol esters derived from free cholesterol (Wang et al., 2014). Liver X receptors (LXRs) are nuclear oxysterol receptors and metabolic sensors, which are initially found to regulate cholesterol metabolism, chronic inflammatory disorders, gluconeogenic pathway and lipid biosynthesis (Geyeregger et al., 2006). LXRα (NR1H3) and LXRβ (NR1H2) are encoded in separate chromosomes sharing more than 75% sequence similarities in their DNA binding domain (DBD) and ligand binding domain (LBD) both in humans and rodents (A-González and Castrillo, 2011). LXRα expression predominates in metabolic tissues such as the liver, intestine, adipose tissue, kidney, and macrophages, whereas LXRβ is more ubiquitously expressed (Mukherjee and Mani, 2010). The first known direct target gene for LXRs in mice was Cyp7a1. LXRs control the expression of Cyp7a1, which promotes cholesterol elimination through the bile. In intestinal epithelial cells, LXRs might increase ABCG5, ABCG8, and ABCA1 expression, thus reducing the intestinal absorption of dietary cholesterol. Similarly, LXRs have also been implicated in fatty acid biosynthesis by inducing the expression of sterol regulatory element-binding protein-1c gene (SREBP-1c) (Repa et al., 2000).
It has been proven that high-glucose and high-fat diet results in higher serum TG, TC and liver TG content as well as increased LXRα mRNA expression, with liver cirrhosis significantly exacerbated in the rat. The Zhengqing Recipe might alleviate liver cirrhosis through the down-regulated expression of LXRα in non-alcoholic fatty liver (Luo et al., 2009). Also, deposition of lipid in hepatocytes might be related to high mRNA expression of LXRα. Panax notoginseng saponins decreased the content of TC in hyperlipidemic hepatocytes and promoted the recovery of hepatocyte steatosis by down-regulating LXRα mRNA (Cheng et al., 2010).
In the present study, when compared with the normal control group, mRNA expression of LXRα in normal ASE groups decreased significantly, and ASE could dose-dependently reduce mRNA expression of LXRα in normal BRL cells. mRNA expression of LXRα in normal BRL cells was low, but it was significantly up-regulated in hyperlipidemic cells. Up-regulated LXRα could regulate target genes involved in TG, unsaturated fatty acids and phospholipids biosynthesis, causing lipid accumulation and hepatocyte steatosis in hyperlipidemic cells. Treatment of mice with synthetic LXR agonists enhanced hepatic TG and increased hepatic lipogenesis has been chiefly attributed to direct induction of SREBP-1c expression by LXRs (Repa et al., 2000). However, after treatment with ASE, LXRα mRNA expression decreased in the hyperlipidemic ASE groups. Thus, ASE reduced mRNA expression of LXRα both in normal and hyperlipidemic BRL cells, suggesting that ASE could inhibit hepatocyte steatosis by down-regulating LXRα.
4.4 Effects of ASE on mRNA expression of FXR in BRL cells
FXR belongs to the superfamily of ligand-activated transcription factors and has the typical nuclear receptor (NR) structure. FXR was identified as a bile acid receptor and biological sensor for the regulation of bile acid biosynthesis and was shown to regulate cholesterol metabolism (Goodwin et al., 2000). Thus FXR constitutes a potential therapeutic target that can be modulated to enhance the removal of cholesterol from the body (Tu et al., 2000; Pineda Torra et al., 2003). When bound to bile acids, FXR repressed transcription of the gene encoding CYP7A1, which was the rate-limiting enzyme in bile acid synthesis, and activated the gene encoding intestinal bile acid-binding protein, which was a candidate bile acid transporter (Makishima et al., 1999). FXR has been shown to be a critical regulator of normal cholesterol metabolism and genetic changes affecting FXR function have the potential to be pro-atherogenic (Lambert et al., 2003). In our study, mRNA expression of FXR was down-regulated by ASE administration both in normal and hyperlipidemic BRL cells, which indicated that ASE might modulate cholesterol metabolism by down-regulation of FXR. It was possible that ASE was a kind of agent that decreased the expression of FXR, thus increasing the mRNA expression of Cyp7a1 and as a result, enhancing the catabolic pathway of cholesterol. Our previous studies have shown that ASE could up-regulate the mRNA expression of Cyp7a1 in rats and hens (Shi et al., 2014; Zhou et al., 2014). The findings also suggested that FXR could be a potential therapeutic target in anticholesterolemic effects induced by ASE. Further research is needed to explain the specific mechanism.
5 Conclusions
Our present study indicated that ASE could up-regulate mRNA expression of Ldlr and down-regulate mRNA expression of LXRα and FXR, thus ameliorating hepatic steatosis. These data indicated that ASE has great potential usefulness as a natural agent for preventing and treating hyperlipidemia.
Compliance with ethics guidelines
Xin-ping LIANG, Dong-qiang ZHANG, Yan-yan CHEN, Rui GUO, Jie WANG, Cheng-zhang WANG, and Ying-hua SHI declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
The procedures of the experiment were strictly according to generally accepted international rules and regulations.
References
Abidi, P., Chen, W., Kraemer, F.B., et al., 2006. The medicinalplant goldenseal is a natural LDL-lowering agent withmultiple bioactive components and new action mechanisms. J. Lipid Res., 47(10):2134–2147. [doi:10.1194/jlr.M600195-JLR200]
A-González, N., Castrillo, A., 2011. Liver X receptors asregulators of macrophage inflammatory and metabolicpathways. BBA-Mol. Basis Dis., 1812(8):982–994. [doi:10.1016/j.bbadis.2010.12.015]
Al-Bar, A., Ismail, A., Cheeke, P.R., et al., 1993. Effect ofdietary Yucca schidigera extract (deodorase) on environmentalammonia and growth performance of chickensand rabbits. Proc. West. Sec. Am. Soc. Anim. Sci., 44:106–108.
Al-Naqeb, G., Ismail, M., Bagalkotkar, G., et al., 2010. Vanillinrich fraction regulates LDLR and HMGCR geneexpression in HepG2 cells. Food Res. Int., 43(10):2437–2443. [doi:10.1016/j.foodres.2010.09.015]
Bittner, V., 2003. Non-high-density lipoprotein cholesteroland cardiovascular disease. Curr. Opin. Lipidol., 14(4):367–371. [doi:10.1097/00041433-200308000-00005]
Brown, M.S., Goldstein, J.L., 1983. Lipoprotein receptors inthe liver. Control signals for plasma cholesterol traffic. J. Clin. Invest., 72(3):743–747. [doi:10.1172/JCI111044]
Brown, M.S., Goldstein, J.L., 1986. A receptor-mediatedpathway for cholesterol homeostasis. Science, 232(4746):34–47. [doi:10.1126/science.3513311]
Cheng, S.B., Yang, Q.H., Zhang, Y.P., et al., 2010. Effects ofpanax notoginseng saponins on content of triglycerideand the mRNA expression of LXRa in steatotic hepatocyteLO2. Chin. J. Pathophysiol., 26(6):1151–1155 (inChinese).
Evangelho, J.S., Casali, K.R., Campos, C., et al., 2011.Hypercholesterolemia magnitude increases sympatheticmodulation and coagulation in LDLr knockout mice. Auton. Neurosci., 159(1-2):98–103. [doi:10.1016/j.autneu.2010.07.010]
Francis, G., Makkar, H.P., Becker, K., 2001. Effects of Quillajasaponins on growth, metabolism, egg production andmuscle cholesterol in individually reared Nile tilapia(Oreochromis niloticus). Comp. Biochem. Physiol. C:Toxicol. Pharmacol., 129(2):105–114. [doi:10.1016/S1532-0456(01)00189-2]
Geyeregger, R., Zeyda, M., Stulnig, T.M., 2006. Liver Xreceptors in cardiovascular and metabolic disease. Cell. Mol. Life Sci., 63(5):524–539. [doi:10.1007/s00018-005-5398-3]
Goodwin, B., Jones, S.A., Price, R.R., et al., 2000. A regulatorycascade of the nuclear receptors FXR, SHP-1, andLRH-1 represses bile acid biosynthesis. Mol. Cell., 6(3):517–526. [doi:10.1016/S1097-2765(00)00051-4]
Ji, W., Gong, B.Q., 2007. Hypolipidemic effects and mechanismsof Panax notoginseng on lipid profile in hyperlipidemicrats. J. Ethnopharmacol., 113(2):318–324.[doi:10.1016/j.jep.2007.06.022]
Kathiresan, S., Manning, A.K., Demissie, S., et al., 2007. Agenome-wide association study for blood lipid phenotypesin the Framingham Heart Study. BMC Med. Genet., 8(Suppl. 1):S17. [doi:10.1186/1471-2350-8-S1-S17]
Kong, W., Wei, J., Abidi, P., et al., 2004. Berberine is a novelcholesterol-lowering drug working through a uniquemechanism distinct from statins. Nat. Med., 10(12):1344–1351. [doi:10.1038/nm1135]
Lambert, G., Amar, M.J., Guo, G., et al., 2003. The farnesoidX-receptor is an essential regulator of cholesterol homeostasis. J. Biol. Chem., 278(4):2563–2570. [doi:10.1074/jbc.M209525200]
Li, Y.H., Yang, P., Kong, W.J., et al., 2009. Berberine analoguesas a novel class of the low-density-lipoproteinreceptor up-regulators: synthesis, structure-activity relationships,and cholesterol-lowering efficacy. J. Med. Chem., 52(2):492–501. [doi:10.1021/jm801157z]
Livak, K.J., Schmittgen, T.D., 2001. Analysis of relative geneexpression data using real-time quantitative PCR and the2-Δ0394;CT method. Methods, 25(4):402–408. [doi:10.1006/meth.2001.1262]
Lopez, A.D., Mathers, C.D., Ezzati, M., et al., 2006. Globaland regional burden of disease and risk factors, 2001:systematic analysis of population health data. Lancet,367(9524):1747–1757. [doi:10.1016/S0140-6736(06)68770-9]
Luo, Q., Wen, X.Y., Lu, M., et al., 2009. Regulation ofZhenqing Recipe on expression of hepatic LXRa in type 2diabetic rats complicated with non-alcoholic fatty liverdisease. World Chin. J. Digestol., 17(14):1394–1401 (inChinese).
Makishima, M., Okamoto, A.Y., Repa, J.J., et al., 1999. Identification of a nuclear receptor for bile acids. Science,284(5418):1362–1365. [doi:10.1126/science.284.5418.1362]
Malinow, M.R., Mclaughlin, P., Stafford, S., et al., 1980.Alfalfa saponins and alfalfa seeds. Dietary effects incholesterol-fed rabbits. Atherosclerosis, 37(3):433–438.[doi:10.1016/0021-9150(80)90148-3]
Mallnow, M.R., Mclaughlin, P., Kohler, G.O., et al., 1977.Prevention of elevated cholesterolemia in monkeys byalfalfa saponins. Steroids, 29(1):105–110. [doi:10.1016/0039-128X(77)90113-1]
Mbikay, M., Sirois, F., Simoes, S., et al., 2014. Quercetin-3-glucoside increases low-density lipoprotein receptor(LDLR) expression, attenuates proprotein convertasesubtilisin/kexin 9 (PCSK9) secretion, and stimulates LDLuptake by Huh7 human hepatocytes in culture. FEBS Open Bio, 4:755–762. [doi:10.1016/j.fob.2014.08.003]
Mukherjee, S., Mani, S., 2010. Orphan nuclear receptors astargets for drug development. Pharm. Res., 27(8):1439–1468. [doi:10.1007/s11095-010-0117-7]
Pineda Torra, I., Claudel, T., Duval, C., et al., 2003. Bile acidsinduce the expression of the human peroxisome proliferatoractivatedreceptor a gene via activation of the farnesoid Xreceptor. Mol. Endocrinol., 17(2):259–272. [doi:10.1210/me.2002-0120]
Pischon, T., Girman, C.J., Sacks, F.M., et al., 2005. Non-highdensitylipoprotein cholesterol and apolipoprotein B inthe prediction of coronary heart disease in men. Circulation,112(22):3375–3383. [doi:10.1161/CIRCULATIONAHA.104.532499]
amírez-Zacarías, J.L., Castro-Muñ ozledo, F., Kuri-Harcuch, W., 1992. Quantitation of adipose conversion and triglyceridesby staining intracytoplasmic lipids with Oil redO. Histochemistry, 97(6):493–497. [doi:10.1007/BF00316069]
Reena, M.B., Gowda, L.R., Lokesh, B.R., 2011. Enhancedhypocholesterolemic effects of interesterified oils aremediated by upregulating LDL receptor and cholesterol7-a-hydroxylase gene expression in rats. J. Nutr., 141(1):24–30. [doi:10.3945/jn.110.127027]
Repa, J.J., Liang, G., Ou, J., et al., 2000. Regulation of mousesterol regulatory element-binding protein-1c gene(SREBP-1c) by oxysterol receptors, LXRa and LXRß. Genes Dev., 14(22):2819–2830. [doi:10.1101/gad.844900]
Rudling, M., 1992. Hepatic mRNA levels for the LDL receptorand HMG-CoA reductase show coordinate regulation invivo. J. Lipid Res., 33(4):493–501.
Santoso, B., Mwenya, B., Sar, C., et al., 2004. Effects of supplementinggalacto-oligosaccharides, Yucca schidigera ornisin on rumen methanogenesis, nitrogen and energymetabolism in sheep. Livest. Prod. Sci., 91(3):209–217.[doi:10.1016/j.livprodsci.2004.08.004]
Sen, S., Makkar, H.P., Becker, K., 1998. Alfalfa saponins andtheir implication in animal nutrition. J. Agric. FoodChem., 46(1):131–140. [doi:10.1021/jf970389i]
Shi, Y.H., Guo, R., Wang, X.K., et al., 2014. The regulation ofalfalfa saponin extract on key genes involved in hepaticcholesterol metabolism in hyperlipidemic rats. PLoS ONE, 9(2):e88282. [doi:10.1371/journal.pone.0088282]
Stockert, J.C., Blá zquez-Castro, A., Cañ ete, M., et al., 2012.MTT assay for cell viability: intracellular localization ofthe formazan product is in lipid droplets. Acta Histochem.,114(8):785–796. [doi:10.1016/j.acthis.2012.01.006]
Tu, H., Okamoto, A.Y., Shan, B., 2000. FXR, a bile acidreceptor and biological sensor. Trends Cardiovasc. Med.,10(1):30–35. [doi:10.1016/S1050-1738(00)00043-8]
Wang, J.M., Wang, D., Tan, Y.Y., et al., 2014. 22(R)-hydroxycholesterol and pioglitazone synergistically decreasecholesterol ester via the PPAR?-LXRa-ABCA1 pathway in cholesterosis of the gallbladder. Biochem. Biophys. Res. Commun., 447(1):152–157. [doi:10.1016/j.bbrc.2014.03.130]
Wang, Y.H., 2007. Effects of alfalfa saponins and alfalfa mealon the production performance and regulation mechanismof weaned piglets and finishing pigs. MS Thesis, Collegeof Animal Science and Veterinary Medicine, Henan AgriculturalUniversity, Zhengzhou, China (in Chinese).
Zhou, L., Shi, Y.H., Guo, R., et al., 2014. Digital geneexpression profiling analysis of the cholesterol-loweringeffects of alfalfa saponin extract on laying hens. PLoSONE, 9(6):e98578. [doi:10.1371/journal.pone.0098578]
Author information
Authors and Affiliations
Corresponding author
Additional information
The two authors contributed equally to this work
Project supported by the National Natural Science Foundation of China (No. 31301983) and the Earmarked Fund for Modern Agro-industry Technology Research System of China (No. CARS-35)
ORCID: Xin-ping LIANG, http://orcid.org/0000-0002-8886-8992
Rights and permissions
About this article
Cite this article
Liang, Xp., Zhang, Dq., Chen, Yy. et al. Effects of alfalfa saponin extract on mRNA expression of Ldlr, LXRα, and FXR in BRL cells. J. Zhejiang Univ. Sci. B 16, 479–486 (2015). https://doi.org/10.1631/jzus.B1400343
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1400343