Abstract
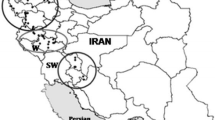
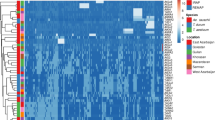
Iran, as a part of Fertile Crescent, is within the center of diversity and origin of the Triticum L. from Poaceae Barnhart (Gramineae A. L. de Jussieu) family. This study concerns the genetic diversity and inter-relationships analyses of 89 accessions of diploid Triticum (2n = 2x = 14) species including T. monococcum, T. boeoticum subsp. boeoticum, T. boeoticum subsp. thaoudar and T. urartu belong to the einkorn wheat group collected all around Iran using 8 primer pairs of IRAP (Inter-Retrotransposon Amplified Polymorphism). Of 220 DNA fragments analysed from all of accessions, 217 (98.6%) were polymorphic. The genetic distances among taxa and accessions were estimated and dendrograms showing relationship among accessions were generated. The results of this study showed that four taxa of diploid wheat are very similar to each other genetically and there is a high affinity, gene flow and genetic relationships between diploid Triticum accessions, although T. monococcum can be distinct loosely from the two other species including T. boeoticum and T. urartu. The highest average Nei genetic distances were found between T. monococcum and T. urartu (0.14) and the lowest between T. boeoticum subsp. boeoticum and T. boeoticum subsp. thaoudar (0.03). Analyssi of molecular variance (AMOVA) was revealed that 84% of total variation attributed to within population’s variability and the remaining 16% variation was ascribed to the differences among species; also it demonstrated a geographical distinction among Triticum accessions. These data proved a center of high diversity in the West and the Northwest of Iran and clearly exposed patterns of two distinct geographic regions.
Similar content being viewed by others
References
Alnaddaf L. M., Moualla M.Y. & Haider N. 2012. Resolving genetic relationships among Aegilops L. and Triticum L. species using analysis of chloroplast DNA by Cleaved Amplified Polymorphic Sequence (CAPS). As. J. Agricul. Scie. 4: 270–279.
Bossolini E., Krattinger S.G. & Keller B. 2006. Development of simple sequence repeat markers specific for the Lr34 resistance region of wheat using sequence information from rice and Aegilops tauschii. Theor. Appl. Genet. 113: 1049–1062.
Boyko E., Kalendar R., Korzun V., Gill B. & Schulman A.H. 2002. Combined mapping of Aegilops tauschii by retrotransposon, microsatellite, and gene markers. Plant Mol. Biol. 48: 767–790.
Cao Q., Lu B.R., Xia H., Rong J., Sala F., Spada A. & Grassi F. 2006. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in north-eastern China revealed by simple sequence repeat (SSR) markers. Ann. Bot. 98: 1241–1252.
Cheniany M., Ebrahimzadeh H., Salimi A. & Niknam V. 2007. Isozyme variation in some populations of wild diploid wheats in Iran. Biochem. Syst. Ecol. 35: 363–371.
Ciaffi M., Dominici L., Umana E., Tanzarella O.A. & Porceddu E. 2000. Restriction fragment length polymorphism (RFLP) for protein disulfide isomerase (PDI) gene sequences in Triticum and Aegilops species. Theor Appl Genet. 101: 220–226.
Dvorak J., Terlizzi P. D., Zhang H.B. & Resta P. 1993. The evolution of polyploid wheats: identification of the A genome donor species. Genome 36: 21–31.
Ehtemam M.H., Rahiminejad M.R., Saeidi H., Ebrahim B., Tabatabaei S., Krattinger S.G. & Keller B. 2010. Relationships among the genomes of Triticum L. species as evidenced by SSR markers in Iran. Int. J. Mol. Sci. 11: 1422–0067.
Figliuolo G. & Perrino P. 2004. Genetic diversity and intraspecific phylogeny of Triticum turgidum L. subsp. dicoccon (Schrank) Thell. revealed by RFLPs and SSRs. Genet. Resour. Crop Evol. 51: 519–527.
Gawel N.J. & Jarret R.L. 1991. A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol Biol Rep. 9: 262–266.
Golovnina K.A., Glushkov S.A., Blinov A.G., Mayorov V.I., Adkison L.R. & Goncharov N.P. 2007. Molecular phylogeny of the genus Triticum L. Plant Syst. Evol. 264: 195–216.
Goncharov N.P., Glushkov S.A. & Shumny V.K. 2008. Domestication of cereal crops in Old World: in search of a new approach to solving old problem. Zhournal Obschei Biologii 68: 125–147.
Harlan J.R. & Zohary D. 1996. Distribution of wild wheats and barely. Sciences 153: 1074–1080.
Heslop-Harrison J.S., Brandes A., Taketa S., Schmidt T., Ver-shinin A.V. & Alkhimova E.G. 1997. The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica 100: 197–204.
Huang S., Sirikhachornkit A. Su X., Faris J., Gill B., Haselkorn R. & Gornicki P. 2002. Genes encoding plastid acetyl-CoA carboxylase and 3- phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploidy wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138.
Kalendar R., Grab T., Regina M., Suoniemi A. & Schulman A.H. 1999. IRAP and REMAP: two new retrotransposon-based DMA fingerprinting techniques. Theor. Appl. Genetics 98: 704–711.
Kalendar R., Tanskanen J., Immonen S., Nevo E. & Schulman A. H. 2000. Genome evolution of wild barely (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 97: 6603–6607.
Kimber G. 1974. A reassessment of the origin of polyploid wheats. Genetics. 78: 487–492.
Kubis S., Schmidt T. & Heslop-Harrison J.S. 1998. Repetitive DNA elements as a major component of plant genomes. Ann. Bot. 82(Supp. A): 45–55.
Li W., Zhang P., Fellers J.P., Friebe B. & Gill B.S. 2004. Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 40: 500–511.
Liu K. & Muse S.V. 2005. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21: 2128–2129.
Malaki M., Naghavi M.R., Alizadeh H., Potki P., Kazemi M., Pirseyedi S. M., Mardi M. & Fakhre-Tabatabaei 2006. Study of genetic variation in wild diploid wheat (Triticum boeoticum) from Iran using AFLP markers. Iran. J. Biotech. 4: 269–274.
Mandy G. 1970. New concept of the origin of Triticum aestivum. Acta Agron. Hung. 19: 413–417.
Manninen O., Kalendar R., Robinson J. & Schulman A.H. 2000. Application of BAR/M retrotransposon markers to the mapping of a major resistance gene for net blotch in barely. Mol. Gener. Gen. 264: 325–334.
Mantel N.A. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27: 209–220 (cited and implemented in NTSYSpc version 02e).
Mizumoto K., Hirosawa S., Nakamura C. & Takumi S. 2002. Nuclear and chloroplast genome genetic diversity in the wild einkorn wheat, Triticum urartu, revealed by AFLP and SSLP analyses. Hereditas 137: 208–214.
Moghaddam M., Ehdaie B. & Waines G. 2000. Genetic diversity in populations of wild diploid wheat Triticum urartu Thum. ex. Gandil. revealed by isozyme markers. Genet. Res. Crop Evol. 47: 323–334.
Morrell P.L., Lundy K.E. & Clegg M.T. 2003. Distinct geographic patterns of genetic diversity are maintained in wild barley (Hordeum vulgare ssp. spontaneum) despite migration. Proc. Natl. Acad. Sci. USA. 100: 10812–10817.
Nei M. & Takezaki N. 1983. Estimation of genetic distances and phylogenetic trees from DNA analysis. In: Proceedings of the 5th World Congress on Genetics applied livestock production 21: 405–412. Cited and implemented in PowerMarker Version 3.0 www.powermarker.com
Nishikawa K., Furuta Y. & Wada T. 1980. Genetic studies on alpha-amylase isozymes in wheat. III. Intraspecific variation in Aegilops squarrosa and the birthplace of hexaploid wheat. Jpn. J. Genet. 55: 325–336.
Olsen K.M. & Purugganan M.D. 2002. Molecular evolution on the origin and evolution of glutinous rice. Genetics 162: 941–950.
Ovesna J., Kucera L., Bockova R. & Holubec, V. 2002. Characterisation of powdery mildew resistance donors within Triticum boeoticum accessions using RAPDs. Czech J Genet. Plant Breed. 38: 117–124.
Provan J., Wolters P., Caldwell K.H. & Powell W. 2004. High-resolution organellar genome analysis of Triticum and Aegilops sheds new light on cytoplasm evolution in wheat. Theor. Appl. Genet. 108: 1182–1190.
Rahiminejad M.R. & Kharazian N. 2005. Illustration of and a key to the genus Triticum L. in Iran. Wheat Information Service 99: 29–34.
Röder M.S, Korzun V., Wendehake K., Plaschke J., Tixier M.H., Leroy P. & Ganal M.W. 1998. A microsatellite map of wheat. Genetics 149: 2007–2023.
Saeidi H., Rahiminejad M.R. & Heslop-Harrison J.S. 2008. Retroelement Insertional Polymorphisms, diversity and phylogeography within diploid, D-genome Aegilops tauschii (Triticeae, Poaceae) Sub-taxa in Iran. Ann. Bot. 101: 855–861.
Sasanuma T., Chabane K., Endo T.R. & Valkoun J. 2002. Genetic diversity of wheat wild relatives in the Near East detected by AFLP. Euphytica 127: 81–93.
Singh, M., Chabane, K., Valkoun, J., Blake, T. 2006. Optimum sample size for estimating gene diversity in wild wheat using AFLP markers. Genet. Res. Crop Evol. 53: 23–33.
Takumi S., Nasuda S., Liu Y.G & Tsunewaki K. 1993. Wheat phylogeny determined by RFLP analysis of nuclear DNA. 1. Einkorn wheat. Jpn J Genet. 68: 73–79.
Teo C.H., Tan S.H., Ho C.L., Faridah Q.Z., Othman Y.R., Heslop-Harrison J.S., Kalendar R. & Schulman A.H. 2005. Genome constitution and classification using retrotransposon-based markers in the orphan crop banana. J. Plant Biol. 48: 96–105.
Van Slageren M.W. 1994. Wild Wheats: Amonograph of Aegilops L. and Amoblyopyrum (Jaoub. & Spach) Eig (Poaceae). Agricultural Univ. Wageningen, Netherlands, pp. 79–104.
Zohary D. & Hopf M. 1993. Domestication of plants in the Old World: the origin and spread of cultivated plants in west Asia, Europe, and Nile valley. Clarendon Press, Oxford, UK, pp. 143–150.
Acknowledgements
The authors would like to thanks from University of Isfahan for its assistance during the project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Eslami Farouji, A., Khodayari, H., Saeidi, H. et al. Genetic diversity of diploid Triticum species in Iran assessed using inter-retroelement amplified polymorphisms (IRAP) markers. Biologia 70, 52–60 (2015). https://doi.org/10.1515/biolog-2015-0002
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1515/biolog-2015-0002