Abstract
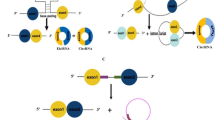
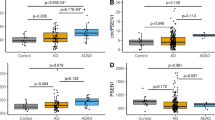
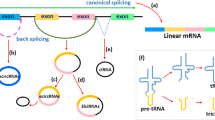
Alzheimer’s disease (AD) is a neurodegenerative disease and there is by far no effective treatment for it, especially in its late stage. Circular RNAs (circRNAs), known as a class of non-coding RNAs are widely observed in eukaryotic transcriptomes, and are reported to play an important role in neurodegenerative diseases including AD. circRNAs usually act as microRNA (miRNA) inhibitors or «sponges» to regulate the function of miRNAs, leading to subsequent changes in protein activities and functions. Accumulating evidence indicates that circRNAs can serve as potential biomarker in AD early prediction. The functional roles of circRNAs are very versatile including miRNAs binding - thus affecting downstream gene expression, generating abnormally translated protein peptides, and affecting epigenetic modifications which subsequently affect AD related gene expressions. Therefore, identifying AD-related circRNAs can contribute to AD early diagnosis and intervention. In this work, we collected and curated an AD-related circRNA dataset; by exploring the circRNAs’ corresponding DNA loci distribution in chromatin 3D conformation (3D genome) and utilize the such 3D genome information, we were able to selected a concise yet predictively effective circRNA panel, based on which, significantly better AD prediction machine learning models were achieved.




Similar content being viewed by others
Availability of data and materials: Data and materials are available up on reasonable request via corresponding authors.
References
Sonkusare SK, Kaul CL, Ramarao P. Dementia of Alzheimer’s disease and other neurodegenerative disorders—memantine, a new hope. Pharmacol Res. 2005;51(1):1–17. https://doi.org/10.1016/j.phrs.2004.05.005
Abeysinghe A, Deshapriya R, Udawatte C. Alzheimer’s disease; a review of the pathophysiological basis and therapeutic interventions. Life Sci. 2020;256:117996. https://doi.org/10.1016/j.lfs.2020.117996
Breijyeh Z, Karaman R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules. 2020;25(24). https://doi.org/10.3390/molecules25245789
Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430(7000):631–9. https://doi.org/10.1038/nature02621
Wang J, Gu BJ, Masters CL, Wang YJ. A systemic view of Alzheimer disease - insights from amyloid-beta metabolism beyond the brain. Nat Rev Neurol. 2017;13(11):703. https://doi.org/10.1038/nrneurol.2017.111
Pereira C, Agostinho P, Moreira PI, Cardoso SM, Oliveira CR. Alzheimer’s disease-associated neurotoxic mechanisms and neuroprotective strategies. Curr Drug Targets CNS Neurol Disord. 2005;4(4):383–403. https://doi.org/10.2174/1568007054546117
Naseri NN, Wang H, Guo J, Sharma M, Luo W. The complexity of tau in Alzheimer’s disease. Neurosci Lett. 2019;705:183–94. https://doi.org/10.1016/j.neulet.2019.04.022
Cummings JL, Tong G, Ballard C. Treatment Combinations for Alzheimer’s Disease: Current and Future Pharmacotherapy Options. J Alzheimers Dis. 2019;67(3):779–94. https://doi.org/10.3233/JAD-180766
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids Are Single-Stranded Covalently Closed Circular Rna Molecules Existing as Highly Base-Paired Rod-Like Structures. P Natl Acad Sci USA. 1976;73(11):3852–6. https://doi.org/10.1073/pnas.73.11.3852
You XT, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603-+.https://doi.org/10.1038/nn.3975
Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58(5):870–85. https://doi.org/10.1016/j.molcel.2015.03.027
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014;9(5):1966–80. https://doi.org/10.1016/j.celrep.2014.10.062
Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep-Uk. 2016;6. https://doi.org/10.1038/srep38907
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. https://doi.org/10.1038/nature11993
Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol. 2020;22(12):2162–9. https://doi.org/10.1007/s12094-020-02371-1
Akhter R. Circular RNA and Alzheimer’s Disease. Adv Exp Med Biol. 2018;1087:239–43. https://doi.org/10.1007/978-981-13-1426-1_19
Cogswell JP, Ward J, Taylor IA, Waters M, Shi YL, Cannon B, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. https://doi.org/10.3233/jad-2008-14103
Lukiw W, Zhao YH, Rogaev E, Bhattacharjee S. A Circular RNA (circRNA) ciRS-7 in Alzheimer’s disease (AD) targets miRNA-7 trafficking and promotes deficits in the expression of the ubiquitin conjugase (UBE2A) and the epidermal growth factor receptor (EGFR). Faseb J. 2016;30. https://doi.org/10.1096/fasebj.30.1_supplement.587.1
Starr DA, Fridolfsson HN. Interactions Between Nuclei and the Cytoskeleton Are Mediated by SUN-KASH Nuclear-Envelope Bridges. Annu Rev Cell Dev Bi. 2010;26:421–44. https://doi.org/10.1146/annurev-cellbio-100109-104037
Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. P Natl Acad Sci USA. 1997;94(3):849–54. https://doi.org/10.1073/pnas.94.3.849
Shin H, Shi Y, Dai C, Tjong H, Gong K, Alber F, et al. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res. 2016;44(7):e70. https://doi.org/10.1093/nar/gkv1505
Mohanta TK, Mishra AK, Al-Harrasi A. The 3D Genome: From Structure to Function. Int J Mol Sci. 2021;22(21). https://doi.org/10.3390/ijms222111585
Ubelmesser N, Papantonis A. Technologies to study spatial genome organization: beyond 3C. Brief Funct Genomics. 2019;18(6):395–401. https://doi.org/10.1093/bfgp/elz019
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326(5950):289–93. https://doi.org/10.1126/science.1181369
Kong SY, Zhang YB. Deciphering Hi-C: from 3D genome to function. Cell Biol Toxicol. 2019;35(1):15–32. https://doi.org/10.1007/s10565-018-09456-2
Shi Y, Guo ZH, Su XB, Meng LM, Zhang MX, Sun J, et al. DeepAntigen: a novel method for neoantigen prioritization via 3D genome and deep sparse learning. Bioinformatics. 2020;36(19):4894–901. https://doi.org/10.1093/bioinformatics/btaa596
Meng L, Wang C, Shi Y, Luo Q. Si-C is a method for inferring superresolution intact genome structure from single-cell Hi-C data. Nat Commun. 2021;12(1):4369. https://doi.org/10.1038/s41467-021-24662-z
Yuan YC, Shi Y, Li CY, Kim JM, Cai WD, Han ZG, et al. DeepGene: an advanced cancer type classifier based on deep learning and somatic point mutations. Bmc Bioinformatics. 2016;17. https://doi.org/10.1186/s12859-016-1334-9
Wu YT, Zhang CJ, Mol B, Kawai A, Li C, Chen L, et al. Early Prediction of Gestational Diabetes Mellitus in the Chinese Population via Advanced Machine Learning. J Clin Endocr Metab. 2021;106(3):E1191–E205. https://doi.org/10.1210/clinem/dgaa899
Wang YF, Shi Y, Zhang CJ, Su KZ, Hu YX, Chen L, et al. Fetal weight estimation based on deep neural network: a retrospective observational study. Bmc Pregnancy Childb. 2023;23(1). https://doi.org/10.1186/s12884-023-05819-8
Yuan YC, Shi Y, Su XB, Zou X, Luo Q, Feng DD, et al. Cancer type prediction based on copy number aberration and chromatin 3D structure with convolutional neural networks. Bmc Genomics. 2018;19. https://doi.org/10.1186/s12864-018-4919-z
Wang MH, Beckmann ND, Roussos P, Wang EM, Zhou XX, Wang Q, et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci Data. 2018;5. https://doi.org/10.1038/sdata.2018.185
Morris JC. Clinical dementia rating: a reliable and valid diagnostic and staging measure for dementia of the Alzheimer type. Int Psychogeriatr. 1997;9 Suppl 1:173–6; discussion 7–8. https://doi.org/10.1017/s1041610297004870
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–87. https://doi.org/10.1101/gr.202895.115
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4). https://doi.org/10.1186/gb-2013-14-4-r36
Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20(11):1666–70. https://doi.org/10.1261/rna.043687.113
Lo I, Hill J, Vilhjalmsson BJ, Kjems J. Linking the association between circRNAs and Alzheimer’s disease progression by multi-tissue circular RNA characterization. Rna Biol. 2020;17(12):1789–97. https://doi.org/10.1080/15476286.2020.1783487
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–80. https://doi.org/10.1038/nature11082
Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–80. https://doi.org/10.1016/j.cell.2014.11.021
Kuhn M. Building Predictive Models in R Using the caret Package. J Stat Softw. 2008;28(5):1–26. https://doi.org/10.18637/jss.v028.i05
Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python (vol 33, pg 219, 2020). Nat Methods. 2020;17(3):352-.https://doi.org/10.1038/s41592-019-0686-2
Hahsler M, Piekenbrock M, Doran D. dbscan: Fast Density-Based Clustering with R. J Stat Softw. 2019;91(1):1–30. https://doi.org/10.18637/jss.v091.i01
Shi Y, Zhang MX, Meng LM, Su XN, Shang XY, Guo ZH, et al. A novel neoantigen discovery approach based on chromatin high order conformation. Bmc Med Genomics. 2020;13. https://doi.org/10.1186/s12920-020-0708-z
Akdemir KC, Le VT, Kim JM, Killcoyne S, King DA, Lin YP, et al. Somatic mutation distributions in cancer genomes vary with three-dimensional chromatin structure. Nat Genet. 2020;52(11):1178-+.https://doi.org/10.1038/s41588-020-0708-0
Perkovic MN, Paska AV, Konjevod M, Kouter K, Strac DS, Erjavec GN, et al. Epigenetics of Alzheimer’s Disease. Biomolecules. 2021;11(2). https://doi.org/10.3390/biom11020195
Szulwach KE, Li XK, Smrt RD, Li YJ, Luo YP, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189(1):127–U81. https://doi.org/10.1083/jcb.200908151
Dudekulay DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. Rna Biol. 2016;13(1):34–42. https://doi.org/10.1080/15476286.2015.1128065
Yang Y, Wu JH, Guan HY, Cai JC, Fang LS, Li J, et al. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. Febs Lett. 2012;586(20):3608–12. https://doi.org/10.1016/j.febslet.2012.08.003
Yang H, Wang H, Shang H, Chen X, Yang S, Qu Y, et al. Circular RNA circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels via directly sponging miR-103 in Alzheimer’s disease (vol 18, pg 2197, 2019). Cell Cycle. 2019;18(24):3603-. https://doi.org/10.1080/15384101.2019.1629773
Shi Y, Su XB, He KY, Wu BH, Zhang BY, Han ZG. Chromatin accessibility contributes to simultaneous mutations of cancer genes. Sci Rep-Uk. 2016;6. https://doi.org/10.1038/srep35270
Wu N, Yuan F, Yue SR, Jiang FY, Ren DC, Liu LJ, et al. Effect of exercise and diet intervention in NAFLD and NASH via GAB2 methylation. Cell Biosci. 2021;11(1). https://doi.org/10.1186/s13578-021-00701-6
Su XB, Zhao LN, Shi Y, Zhang R, Long Q, Bai SH, et al. Clonal evolution in liver cancer at single-cell and single-variant resolution. J Hematol Oncol. 2021;14(1). https://doi.org/10.1186/s13045-021-01036-y
Guo ZH, Liu LJ, Feng MF, Su K, Chi RQ, Li KY, et al. 3D genome assisted protein-protein interaction prediction. Future Gener Comp Sy. 2022;137:87–96. https://doi.org/10.1016/j.future.2022.07.005
Chen NF, Zhao G, Yan X, Lv Z, Yin HM, Zhang SL, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19. https://doi.org/10.1186/s13059-018-1594-y
Lee WJ, Moon J, Jeon D, Shin YW, Yoo JS, Park DK, et al. Possible epigenetic regulatory effect of dysregulated circular RNAs in Alzheimer’s disease model. Sci Rep-Uk. 2019;9. https://doi.org/10.1038/s41598-019-48471-z
Li Y, Zheng QP, Bao CY, Li SY, Guo WJ, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4. https://doi.org/10.1038/cr.2015.82
Funding
Funding: This project was supported by the Key Research and Development Plan of the Ministry of Science and Technology (2022YFE0125300), the National Key Research and Development Program (2016YFC0906400), the Innovation Funding in Shanghai (20JC1418600 and 18JC1413100), the National Natural Science Foundation of China (82071262 and 81671326), the Natural Science Foundation of Shanghai (20ZR1427200 and 20511101900), the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), the Shanghai Key Laboratory of Psychotic Disorders (13DZ2260500), the Shanghai Leading Academic Discipline Project (B205), the Shanghai Jiao Tong University STAR Grant (YG2023ZD26, YG2022ZD024, and YG2022QN111), and Shanghai Gaofeng & Gaoyuan Project for University Academic Program Development (Shanghai University of Sport-2023).
Author information
Authors and Affiliations
Contributions
Authors’ contributions: RC, KL, KS and LL participated in the omics and computational experiments. RC and KL conducted the computational experiments. RC provided the figures, tables, and drafted the manuscript. RC, KL and LL designed chromatin modeling. JW, XL and GH provided clinical suggestions and biological insights about AD. YS and GH initiated this project and supervised the whole workflow. YS and GH edited and finalized the manuscript. All the authors reviewed and proof read the manuscript and the experimental results.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate: Not applicable.
Competing interests: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Rights and permissions
About this article
Cite this article
Chi, R., Li, K., Su, K. et al. Prediction of Alzheimer’s Disease Based on 3D Genome Selected circRNA. J Prev Alzheimers Dis (2024). https://doi.org/10.14283/jpad.2024.52
Received:
Accepted:
Published:
DOI: https://doi.org/10.14283/jpad.2024.52