Abstract
In recent years, Generalized Modules for Membrane Antigens (GMMA) have received increased attention as an innovative vaccine platform against bacterial pathogens, particularly attractive for low- and middle-income countries because of manufacturing simplicity. The assessment of critical quality attributes (CQAs), product-process interactions, identification of appropriate in process analytical methods, and process modeling is part of a robust quality by design (QbD) framework to support further development and control of manufacturing processes. QbD implementation in the context of the GMMA platform will ensure robust manufacturing of batches with desired characteristics, facilitating technical transfer to local manufacturers, regulatory approval, and commercialization of vaccines based on this technology. Here, we summarize the methodology suggested, applied to a first step of GMMA manufacturing process.
Graphical Abstract

Similar content being viewed by others
Introduction
Outer membrane vesicles (OMV) are spherical bi‐layered membrane structures with a diameter in the range of 20–250 nm, naturally released by Gram-negative bacteria during their growth (1). They resemble the bacterial surface of the pathogen and are primarily made of bacterial outer membrane constituents, therefore containing key antigenic components to elicit a protective immune response. OMV also contain lipopolysaccharides (LPS) and other pathogen-associated molecular patterns (2), have optimal size for cellular uptake, and display antigens in their native conformation and orientation, representing a promising vaccine platform (3, 4). Indeed, a meningitis serogroup B vaccine, which contains a bacterial OMV component, was approved for human use in 2013 (5), and many other vaccine candidates based on this technology are under development, targeting different diseases (6).
Bacteria can be genetically manipulated to enhance OMV release, resulting in hyperblebbing microorganisms whose blebs have been also called Generalized Modules for Membrane Antigens (GMMA) (7). Additional genetic modifications are generally introduced to reduce LPS reactogenicity (8, 9), most often by modifying the lipid A structure of LPS molecules. GMMA production process is simple and highly cost-effective (10), which makes GMMA particularly attractive for the development of vaccines against neglected diseases for impoverished communities (11, 12). This technology has been applied in fact to different pathogens having a huge burden of disease in low- and middle-income countries (LMICs), like invasive non-typhoidal Salmonella (iNTS) (13), Neisseria meningitidis (14), and Shigella, with the most advanced candidate being tested in phase 2 clinical trials (15,16,17).
Purification of GMMA is straightforward: a first microfiltration (MF) step allows separation of GMMA, secreted in the culture medium, from whole bacteria, while a second ultrafiltration (UF) step retains GMMA while removing soluble proteins and other low molecular weight impurities, allowing also buffer changing (Fig. 1a) (7).
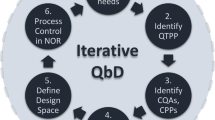
Over the last years, the quality by design (QbD) approach has been implemented to ensure consistent delivery of safe and efficacious products and to provide a framework for prior knowledge establishment and use, in order to enable a faster access to drugs and vaccines. Starting from the definition of quality target product profile (QTPP) that defines intended use of the drug product in clinical setting, administration route, dosage form, and desired quality characteristics of the drug product, QbD relies on deep product and process understanding and on having control strategies in place (18). The application of QbD to technology platforms can allow further improvement of the technology and accelerate the development of novel products based on it (19).
Here, we applied QbD to the GMMA production process. We focus on the downstream process of drug substance manufacturing (Fig. 1a) and selected the UF step as the first focal point. Potentially, critical GMMA quality attributes (to ensure safety and efficacy) were listed, and process parameters of the selected step that could affect final product quality (defined as potential critical process parameters (pCPPs)) were identified based on prior knowledge.
Mathematical models linking critical quality attributes (CQAs) to critical process parameters (CPPs) were built and used to identify the process design space to reduce product variability and increase process capability.
Systematic enhanced knowledge on the product and on the process through QbD should enable efficient scale-up, facilitate technology transfer to local manufacturers, regulatory approval, and commercialization together with post-approval changes and root cause analysis (20,21,22).
S. Typhimurium GMMA were selected as model to be used in this work.
S. Typhimurium is the most common serovar responsible for invasive non-typhoidal Salmonella, a leading cause of death and morbidity in Africa (23). GMMA have been proposed as innovative delivery system for the O-antigen (OAg), recognized as target of protective immunity. S. Typhimurium OAg is constituted by a five-sugar repeating unit with a backbone made of galactose (Gal), rhamnose (Rha), and mannose (Man), and a 3,6-dideoxy-hexose (abequose, Abe) linked to Man conferring O:4 specificity (24). Abe may be O-acetylated at position C-2, which adds the O:5 specificity. OAg chains can also be variably glucosylated, with glucose (Glc) linked to Gal. OAg repeating units are linked to the core oligosaccharide attached to lipid A through a 3-deoxy-d-manno-octulosonic acid (KDO) sugar to form the entire LPS molecule.
S. Typhimurium GMMA used in this study display OAg populations at different average sizes of 30 KDa and 2 KDa (the peak at 2 kDa includes core with no OAg repeats and core with few OAg repeats attached) (24). O-acetylation on Abe is around 60%.
Material and Methods
GMMA Production
GMMA were produced in flasks from a mutant strain of Salmonella enterica serovar Typhimurium isolate 2192 (ΔtolR::aph, ΔmsbB::tet, ΔpagP::cat, LT-2 collection, University of Calgary).
A working cell bank was used to inoculate 200 mL of chemically defined medium (CDM) starting from approximately 0.04 OD/mL in a 1-L baffled flask (1:5 ratio of culture medium to flask volume) and grown overnight (ON) at 30°C with agitation (180 rpm). The day after, 4 × 2-L baffled flasks (each one containing 700 mL of CDM) were inoculated with the ON growth starting from approximately 0.24 OD/mL. After 8 h, bacteria cultures were centrifuged at 4000 rpm for 1 h. Supernatants were collected and sterile filtered.
CDM composition: KH2PO4 13.3 g/L, (NH4)2HPO4 4 g/L, citric acid monohydrate 1.7 g/L, dextrose monohydrate 10 g/L, l-methionine 0.16 g/L, l-threonine 0.22 g/L, l-arginine 0.56 g/L, l-aspartic acid 1 g/L, l-proline 0.4 g/L, l-glutamic acid monosodium salt monohydrate 1.78 g/L, H2SO4 20 µL/L, MnCl2·4 H2O 15 mg/L, CuCl2·2 H2O 1.5 mg/L, H3BO3 3 mg/L, Na2MoO4·2 H2O 2.5 mg/L, zinc acetate dihydrate 13 mg/L, [Co(NH3)6]Cl3 2.5 mg/L, ferric citrate 49 mg/L, MgSO4·7 H2O 493 mg/L, thiamine hydrochloride 50 mg/L, nicotinic acid 10 mg/L, pyridoxine hydrochloride 10 mg/L, calcium pantothenate 10 mg/L, cyanocobalamin 1 mg/L.
Tangential flow filtration (TFF) was performed using a Sartorius Sartoflow system equipped with a SciLog Peristaltic Pump Tandem 1082. The ultrafiltration membrane used was from Sartorius with 300 KDa cutoff with 0.1 m2 area in polyethersulfone.
Analytical Methods
GMMA particle size was determined by dynamic light scattering (DLS) and by high performance size exclusion liquid chromatography (HPLC-SEC) coupled with multi-angle laser light scattering (MALLS); total protein was quantified by micro-BCA assay, while GMMA content, soluble protein, and DNA impurities were determined by HPLC-SEC. For OAg characterization, O-acetyl content was determined by 1H-NMR; quantification was performed by high performance anionic exchange chromatography with pulsed amperometric detection (HPAEC-PAD); HPLC-SEC was used to determine OAg size and lipid A (by quantifying terminal KDO of extracted OAg after semicarbazide derivatization). All methods are detailed in (25).
Statistical analysis was performed with Minitab 18.1.0 (Minitab Inc.) and with Design-Expert 10 (Stat Ease).
QbD Methodology
QbD methodology was applied to the UF step of GMMA Drug Substance manufacturing. For starting the possibility to simplify the original UF, process was evaluated. Potential GMMA CQAs were listed, and the impact of the UF step on them was evaluated. Based on this assessment, only CQAs directly or indirectly impacted by the UF step were considered for establishing a relationship between CQAs and pCPPs. The impact of each pCPP on the CQAs was evaluated by assigning a severity score, and pCPP resulting in the highest score on the total of CQAs was then prioritized and used as factors in the optimization phase of the UF step by design of experiment (DoE). Also, appropriate analytical methods to characterize the intermediate samples coming from the UF step were selected and characterized to verify if appropriate for building statistical models through the DoE approach. A response surface method (RSM) central composite design (CCD) face centered was used. All runs were performed with two replicates of factorial points and four replicates of the central point. A total of 26 runs were performed in randomized order (Table S1). One single bacterial growth was used, and the collected supernatant was aliquoted in 30 different aliquots stored at − 80°C; 1 aliquot was used for each run. By this way, the variability of the starting material was not a parameter to keep in consideration. For each response surface, residuals versus run diagnostics were checked to identify any possible trend due to starting material stability. ANOVA analysis for mathematical model significance was performed for each response. Not significant terms (p-value > 0.05) were removed from the model using a backward elimination process, where appropriate (Box-Cox analysis) data were transformed.
Process optimization was performed with the boundary of the design space for the selected factors (pH starting, TMP, DV), the boundary of the product specification (soluble protein, pH, OAg/protein ratio, O-acetyl content), the goal to minimize the impurities (soluble protein, DNA), and the propagation of error (POE) of all CQAs (Table S2 reports the specification boundaries and the goal in the optimization for each CQA).
Monte Carlo simulation was used to study the effect of CPPs variability on CQAs and perform a process capability analysis. Monte Carlo simulations (n = 150,000) were run considering CPPs normal distributions centered on optimized settings with the variability previously determined.
Results and Discussion
Ultrafiltration Process Analysis
Preliminary Studies to Simplify the Process and Reduce the Number of pCPP
For starting the UF step was analyzed (Fig. 1b), and some tests were planned to verify the possibility to further simplify the process and reduce the number of pCPP.
The UF step consists of two diafiltration steps differing in number of diavolumes and diafiltration buffers:
-
First step: six diavolumes (DV) with PBS
-
Second step: ten DV with saline
Furthermore, processed material is concentrated before and after the first diafiltration step (Fig. 1b).
We evaluated the possibility of simplifying the process, understanding the impact that concentration and diafiltration steps could have on the purification and on the quality of the final product. In particular, we evaluated the possibility of avoiding the use of two different diafiltration buffers (Supporting Information (SI), Run 1 vs Run 2) and eliminating an intermediate concentration step (SI Run 2 vs Run 3).
GMMA coming from these three runs showed similar characteristics, as reported in Table S3 and Figure S1; therefore, the process was simplified as in Run 3 (Fig. 1c): growth supernatant was adjusted to pH 7.2, concentrated directly 12 × , and diafiltered against saline only (16 DV). Moreover, samples collected as intermediates during each purification were analyzed for soluble proteins and DNA impurities to improve our knowledge on the overall process (Table S4).
Selection of Critical Quality Attributes (CQA) to Monitor
Table I lists the potential GMMA CQAs: some of them refer to GMMA as particle; other relate to key antigens presented on GMMA surface, e.g., OAg of LPS molecules. Such list was established on the basis of prior knowledge, current structure–function understanding, strategic nonclinical studies performed with S. Typhimurium GMMA.
We have evaluated if the UF step can have an impact on each the potential CQAs.
Depending on the possible impact, the potential CQAs are classified as impacted (I)/not impacted (NI)/potentially impacted (PI). The classification has been carried out based on product and process understanding know-how.
In Table I, the secondary interactions between potential CQAs, assigned based on logical reasoning and the chemical-physical knowledge of the drug substance, are reported. As an illustrative example, with a change in molecular size distribution of the OAg, a change also in OAg quantification and OAg to protein ratio can be expected.
Definitions of the Process
Based on the assessment reported above, only CQAs directly or indirectly impacted by the UF step were considered for establishing a relationship between CQAs and pCPP (Table IIA). This first assessment was based on current know-how and historical data collected. The impact of each pCPP on each CQA has been classified following the criteria reported in Table IIB, and the type of relationship (if known) has been indicated:
-
ID: if the increase of the pCPP value causes a decrease of CQA value and vice versa (decrease-increase)
-
II: if the increase of the pCPP value causes an increase of CQA value and vice versa (decrease-decrease)
-
Data analysis: if the type of the relationship is not clear and additional data are needed.
GMMA concentration in the material to be processed (coming from the previous purification step) could also have an impact, but it has not been considered at this stage.
Other parameters such as UF membrane cutoff and material, diafiltration buffer composition and raw materials that clearly can have an impact on the CQAs but are considered fixed at this stage of the study as they are already defined in the process and cannot be easily modified.
From the total scores calculated in Table IIA, the most critical process parameters that impact CQAs resulted to be transmembrane pressure (TMP), number of DV, and starting pH.
Measurement System Analysis (MSA)
For the purpose of this study, it was important to identify appropriate analytical methods to characterize the intermediate samples coming from the UF step.
HPLC-SEC was used to determine soluble proteins, DNA impurities, and GMMA yield. Total OAg was estimated by HPAEC-PAD, while total protein content was determined by micro-BCA, and their ratio was calculated. OAg size distribution (molecular weight and OAg to core ratio) and lipid A content were determined by HPLC-SEC/semicarbazide on extracted OAg. Protein pattern was qualitatively investigated by SDS-PAGE as well as appearance. O-acetyl content was quantified by 1H NMR. Two different methods, HPLC-SEC/MALLS and DLS, were used to determine particle size.
It was deemed appropriate to characterize DLS, HPLC-SEC methods, micro-BCA, and HPAEC-PAD for their reproducibility to verify if appropriate for building statistical models through the DoE approach, considering the minimum signal change detectable/noise ratio that has been predicted from the power calculation in DoE planning.
The characterization design was planned with two different operators for a total of six independent analytical sessions (three for each operator). In the analysis of variance, the sessions were nested into the operator as it was not possible for the operators to perform the analysis in parallel.
For each analytical method, samples were prepared in triplicate at multiple concentrations starting from a GMMA standard lot to investigate the performance of the analytical method at different concentration levels (i.e., close to the lowest, the middle and the highest level of the analytical method calibration curve).
For GMMA protein content and purity (soluble protein and nucleic acid) by HPLC-SEC, two samples differing in the content of impurities, to represent also the worst case, were analyzed in triplicate: bacteria culture supernatant (unprocessed material, thus containing a high percentage of soluble proteins and high 260/280 nm Abs ratio) and TFF retentate after 13 DV (not completely purified GMMA, with a certain percentage of soluble proteins), analyzed respectively neat and with a twofold dilution.
For each analysis session, the preparation and the analysis order of the samples were randomized.
Variance component analyses were performed with mixed effects model, and results are reported in Table III (per each analytical method/sample type, statistical analysis is reported in SI Tables S5-S18). The precision of each method is expressed as coefficient of variation (CV), considering the average of all measurements for each concentration tested.
Optimization Phase (DoE)
Design, Factors, and Responses
At this point, the DoE approach was used to investigate the impact of selected pCPP on GMMA CQAs. The following three key factors were used: starting pH, TMP, and number of DV. As mainly quadratic models were expected for the responses under evaluation, RSM design was selected from the beginning without performing a preliminary screening design and then a characterization design.
This choice was made because only three input factors were selected based on the assessment reported in Table IIA.
Parameter ranges were selected on the following basis:
-
Starting pH: suitable range to avoid O-acetyl groups hydrolysis and protein precipitation based on prior knowledge (26) (setting: low 5.3, middle 6.25, high 7.2);
-
TMP: instrumental technical feasibility (setting: low 0.5, middle 0.875, high 1.25 bar);
-
DV: from preliminary study which results are reported in Table S4 (low 5; middle 13, high 21).
A face centered design, instead of a rotatable one, was chosen as the parameters cannot be set far outside the design space due to GMMA product stability (starting pH), instrumental boundaries (TMP), and based on technical knowledge (DV).
With the design described, a quadratic model would be able to detect a signal/noise ratio of 2 standard deviations at 5% alpha level with a power of 97.8% for order 1 term, 96.3% for two-way interactions, and 87.9% for quadratic terms. The related Fraction of Design Space (FDS) graph is reported in Figure S2.
Thirteen output responses among the GMMA CQAs have been evaluated: soluble proteins (1), DNA content (2), GMMA size by HPLC-SEC MALLS (3), GMMA size by DLS (4), GMMA size polydispersity index by DLS (5), pH of product (6), osmolality (7), GMMA yield (8), OAg/total protein ratio (9), O-acetyl content (10), OAg size (11), OAg to core ratio (12), and lipid A to GMMA protein ratio (13). GMMA content (8) was also evaluated to get insight on process yield.
DoE Elaboration
After having obtained models for the different attributes (only for lipid A and DLS Z-average a model was not obtained, Table IV) and identified the dependence between CPP and each of CQA, we concentrated our attention on the quality of the drug substance and not on the yield, to identify the design space to purify GMMA with CQAs inside set boundaries. GMMA yield was investigated, but it was outside the scope of this work that aimed to ensure good quality of the final product and not to improve yields. This is related to the fact that manufacturability of GMMA is simple and at low cost and yields obtained are already satisfactory.
Among all the responses considered, only the following ones were used: soluble proteins, DNA content, pH, OAg/total protein ratio, and O-acetyl content for process optimization. Particle size distribution by MALLS, osmolality, OAg size, and OAg/core ratio gave small variation respect to the boundaries fixed in the entire design space.
Increasing the number of DV, impurities (nucleic acids and soluble proteins) are reduced and consequently the OAg/total protein ratio increased.
Starting pH, as expected, has the major impact in determining the purified GMMA pH (in case of higher starting pH, a higher number of DV contributes to achieve a lower purified GMMA final pH). Starting pH values close to 6.5 resulted the optimal to preserve O-acetylation level that is also negatively impacted by higher number of DV.
By increasing TMP probably, there is an impact on GMMA integrity resulting in an increase of soluble proteins especially with lower DV number.
Variability of the Process Factors
For the process optimization, also the POE was considered in order to find factor settings that minimize variation transmitted to the response from each factor; by this way, the process will be more robust to variations in input factors.
To perform the model optimization with POE, we considered the following variability of the factors:
-
The equipment measures the DV by weighing the TFF permeate and the standard deviation reported for the instrumental weighing system is of 0.1 g. Considering that 1 DV corresponds to 25 g, the standard deviation considered for the DV factor corresponds to 0.004 DV (25 g:1dv = 0.1:x).
-
The TMP is measured with a pressure transducer and the standard deviation reported for the device used is of 0.02 bar.
-
For the starting material pH measurement, the pH meter variability reported in the instrument specification is not deemed appropriate as the sample is very dirty, so it has been measured on real sample. The characterization design was planned with one operator for three independent analytical sessions (one per day); in each analytical session, three different measurements were performed on the same real sample. In Table S32, the variance component analysis, obtained with general linear model, is reported. The standard deviation calculated from the total variance is 0.11 pH unit.
Optimization with POE
Optimal conditions for performing the UF step resulted to be starting pH 6.7, TMP 0.56 bar, and DV 17.3 (the optimization without POE minimization would lead instead to the following conditions: starting pH 7.2, TMP 0.50 bar, DV 18.6). In Table S33, the 95% CI for mean and tolerance interval for the predicted values of CQAs in the optimal conditions identified are reported.
In Figure S36, desirability ramps for each factor and each response, as well as the combined desirability, are reported. In Fig. 2, the combined desirability function calculated in the design space is reported showing the process parameter (PP) ranges to obtain purified GMMA with CQA mean responses in the specifications set. In Figure S37, S38, and S39, the graphs of the CQA models close to the optimized conditions are reported. In Figure S40, S41, and S42, the graphs of the POE for CQA close to the optimized conditions are reported.
Model Confirmation
To confirm the model, an additional UF run with the same starting material was performed using the optimized conditions identified. The TFF retentate was fully characterized (Table S34), and the responses obtained were evaluated respect to the predictions (Table S35): all attributes fell in the predicted ranges.
The study was conducted by ignoring possible variability coming from the starting material, and the runs were performed at small scale. Of course, it will be important to verify that expected results are obtained on material coming from different fermentation runs and by performing the TFF at bigger scale. Working at small scale allowed to execute multiple runs in relatively short period of time and to understand impact that process parameters can have on GMMA quality. By scaling up the process, changes could be expected, but the knowledge acquired will allow to rapidly identify optimal working conditions if needed. It will be interesting to apply conditions identified in this study for S. Typhimurium to GMMA from other pathogens and confirm platform potential of the process. We could expect need for certain adjustments based on specific antigen characteristics, but again, the info acquired will allow to accelerate the work on GMMA from different organisms.
Capability Analysis
We evaluated, in the optimized CPPs settings identified by DoE, the effect of CPPs variabilities on CQAs variabilities.
Monte Carlo simulations were run on CPPs, and, by using the transfer functions found with the DoE, the resulting CQAs distributions were identified (Fig. 3 and Table V) and studied with the capability analysis to understand the probability of an out-of-specification. Results of the capability analysis are reported in Table VI.
For all parameters, except pH and OAg/protein ratio, Ppk is higher than 1.33, meaning that the process is appropriate for meeting the boundaries set. For pH and OAg/protein ratio, Ppk values are close to 1, and in addition, the process is not centered on the target. Therefore, the tolerances for the key factors that influence the process (starting pH, TMP, DV) as well as the MSA values for the CQAs need to be kept under control with time.
The results obtained are considered acceptable for a biological process as the highest probability of out of spec is 0.16%.
Conclusions
In this work, CPPs and CQAs for GMMA have been listed, and their relationship established for a first step of the manufacturing process, identified as critical. S. Typhimurium GMMA, has been used as model. The TFF process has been characterized in the designed space for the attributes selected. A model has been created based on five attributes identified as critical. Optimized conditions have been found to minimize impurities and have pH, OAg/protein ratio, and O-acetyl content in the specification ranges set. One TFF run has been conducted in the optimized conditions as confirmation of the models, and the resulting material had all five parameters inside the predicted ranges. Finally, a Monte Carlo simulation has been performed indicating a low probability of failure.
The exercise performed on the TFF step needs to be extended to other critical steps of the process, e.g., fermentation and microfiltration, but represents clear evidence of how QbD can be applied to vaccine manufacturing processes to guarantee quality and reduce risk of failure. Design of experiment has helped to determine correlation, direction, and magnitude of changes, with the final goal to establish semi-mechanistic models that quantitatively link CPPs to CQAs and to define the design space for GMMA manufacturing.
To our knowledge, no vaccines have yet been developed under a full and integrated QbD framework, but recent progress in analytical and computational capabilities makes feasible the implementation of this approach facilitating the development of transferable production processes under a solid control strategy. This exercise is important to set the stage for process performance qualification and ensure clear understanding of source of variability for future routine manufacturing execution and continued improvements. Indeed, process development under QbD framework is an iterative, as mathematical modeling, analysis, and real-time measurements can be used to reassess parameter criticality and develop an appropriate life cycle management strategy. The acquired knowledge is of particular value when can be transferred from one product/process to another in the context of a platform technology as GMMA.
Data Availability
The authors declare that data are contained within the article and in the Supplementary Materials.
References
Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13(10):605–19. https://doi.org/10.1038/nrmicro3525.
Mancini F, Rossi O, Necchi F, Micoli F. OMV vaccines and the role of TLR agonists in immune response. International journal of molecular sciences. 2020;21(12). https://doi.org/10.3390/ijms21124416.
Li M, Zhou H, Yang C, Wu Y, Zhou X, Liu H, Wang Y. Bacterial outer membrane vesicles as a platform for biomedical applications: an update. J Control Release: Off J Control Release Soc. 2020;323:253–68. https://doi.org/10.1016/j.jconrel.2020.04.031.
van der Pol L, Stork M, van der Ley P. Outer membrane vesicles as platform vaccine technology. Biotechnol J. 2015;10(11):1689–706. https://doi.org/10.1002/biot.201400395.
O’Ryan M, Stoddard J, Toneatto D, Wassil J, Dull PM. A multi-component meningococcal serogroup B vaccine (4CMenB): the clinical development program. Drugs. 2014;74(1):15–30. https://doi.org/10.1007/s40265-013-0155-7.
Micoli F, MacLennan CA. Outer membrane vesicle vaccines. Semin Immunol. 2020;50:101433. https://doi.org/10.1016/j.smim.2020.101433.
Gerke C, Colucci AM, Giannelli C, Sanzone S, Vitali CG, Sollai L, et al. Production of a Shigella sonnei vaccine based on Generalized Modules for Membrane Antigens (GMMA), 1790GAHB. PLoS One. 2015;10(8):e0134478. https://doi.org/10.1371/journal.pone.0134478.
Rossi O, Caboni M, Negrea A, Necchi F, Alfini R, Micoli F, et al. Toll-like receptor activation by Generalized Modules for Membrane Antigens from lipid A mutants of Salmonella enterica serovars Typhimurium and Enteritidis. Clin Vaccine Immunol. 2016;23(4):304–14. https://doi.org/10.1128/CVI.00023-16.
Rossi O, Pesce I, Giannelli C, Aprea S, Caboni M, Citiulo F, et al. Modulation of endotoxicity of Shigella generalized modules for membrane antigens (GMMA) by genetic lipid A modifications: relative activation of TLR4 and TLR2 pathways in different mutants. J Biol Chem. 2014;289(36):24922–35. https://doi.org/10.1074/jbc.M114.566570.
Kis Z, Shattock R, Shah N, Kontoravdi C. Emerging technologies for low-cost, rapid vaccine manufacture. Biotechnol J. 2019;14(1):e1800376. https://doi.org/10.1002/biot.201800376.
Micoli F, Adamo R, Nakakana U. Outer membrane vesicle vaccine platforms. BioDrugs. 2023. https://doi.org/10.1007/s40259-023-00627-0.
Piccioli D, Bartolini E, Micoli F. GMMA as a ‘plug and play’ technology to tackle infectious disease to improve global health: context and perspectives for the future. Expert Rev Vaccines. 2022;21(2):163–72. https://doi.org/10.1080/14760584.2022.2009803.
Micoli F, Rondini S, Alfini R, Lanzilao L, Necchi F, Negrea A, et al. Comparative immunogenicity and efficacy of equivalent outer membrane vesicle and glycoconjugate vaccines against nontyphoidal Salmonella. Proc Natl Acad Sci U S A. 2018;115(41):10428-33.https://doi.org/10.1073/pnas.1807655115.
Koeberling O, Ispasanie E, Hauser J, Rossi O, Pluschke G, Caugant DA, et al. A broadly-protective vaccine against meningococcal disease in sub-Saharan Africa based on generalized modules for membrane antigens (GMMA). Vaccine. 2014;32(23):2688–95. https://doi.org/10.1016/j.vaccine.2014.03.068.
Launay O, Lewis DJM, Anemona A, Loulergue P, Leahy J, Scire AS, et al. Safety profile and immunologic responses of a novel vaccine against Shigella sonnei administered intramuscularly, intradermally and intranasally: results from two parallel randomized phase 1 clinical studies in healthy adult volunteers in Europe. EBioMedicine. 2017;22:164–72. https://doi.org/10.1016/j.ebiom.2017.07.013.
Launay O, Ndiaye AGW, Conti V, Loulergue P, Scire AS, Landre AM, et al. Booster vaccination with GVGH Shigella sonnei 1790GAHB GMMA vaccine compared to single vaccination in unvaccinated healthy European adults: results from a phase 1 clinical trial. Front Immunol. 2019;10:335. https://doi.org/10.3389/fimmu.2019.00335.
Obiero CW, Ndiaye AGW, Scire AS, Kaunyangi BM, Marchetti E, Gone AM, et al. A Phase 2a randomized study to evaluate the safety and immunogenicity of the 1790GAHB Generalized Modules for Membrane Antigen vaccine against Shigella sonnei administered intramuscularly to adults from a Shigellosis-endemic country. Front Immunol. 2017;8:1884. https://doi.org/10.3389/fimmu.2017.01884.
Rathore AS, Winkle H. Quality by design for biopharmaceuticals. Nat Biotechnol. 2009;27(1):26–34. https://doi.org/10.1038/nbt0109-26.
Daniel S, Kis Z, Kontoravdi C, Shah N. Quality by design for enabling RNA platform production processes. Trends Biotechnol. 2022;40(10):1213–28. https://doi.org/10.1016/j.tibtech.2022.03.012.
Zhang L, Mao S. Application of quality by design in the current drug development. Asian J Pharm Sci. 2017;12(1):1–8. https://doi.org/10.1016/j.ajps.2016.07.006.
Rathore AS. Roadmap for implementation of quality by design (QbD) for biotechnology products. Trends Biotechnol. 2009;27(9):546–53. https://doi.org/10.1016/j.tibtech.2009.06.006.
Looby M, Ibarra N, Pierce JJ, Buckley K, O’Donovan E, Heenan M, et al. Application of quality by design principles to the development and technology transfer of a major process improvement for the manufacture of a recombinant protein. Biotechnol Prog. 2011;27(6):1718–29. https://doi.org/10.1002/btpr.672.
Marchello CS, Fiorino F, Pettini E, Crump JA. Incidence of non-typhoidal Salmonella invasive disease: a systematic review and meta-analysis. J Infect. 2021;83(5):523–32. https://doi.org/10.1016/j.jinf.2021.06.029.
De Benedetto G, Alfini R, Cescutti P, Caboni M, Lanzilao L, Necchi F, et al. Characterization of O-antigen delivered by Generalized Modules for Membrane Antigens (GMMA) vaccine candidates against nontyphoidal Salmonella. Vaccine. 2017;35(3):419–26. https://doi.org/10.1016/j.vaccine.2016.11.089.
Micoli F, Alfini R, Giannelli C. Methods for assessment of OMV/GMMA quality and stability. In: Bidmos F, Bossé J, Langford P, editors. Bacterial Vaccines: Methods and Protocols. New York, NY: Springer US; 2022. p. 227–79.
Micoli F, Giannelli C, Di Benedetto R. O-antigen extraction, purification, and chemical conjugation to a carrier protein. Methods Mol Biol (Clifton, NJ). 2021;2183:267–304. https://doi.org/10.1007/978-1-0716-0795-4_14.
Acknowledgements
We thank GVGH and GSK colleagues that contributed to CQA and CPP assessment.
Funding
This research was partially funded by the UK Department of Health and Social Care, grant number EP/R013764/1.
Author information
Authors and Affiliations
Contributions
Conceptualization: CG, FN, MMP, ZK, CK, CC, and FM; performed the experiments: EP, DO, and BR; methodology: all authors; formal analysis: all authors; writing—original draft preparation: CG and FM. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
GSK Vaccines Institute for Global Health Srl is an affiliate of GlaxoSmithKline Biologicals SA. C.G, F.N., E.P., D.O., C.C. and F.M. are employees of the GSK group of companies. B.R. was an employee of the GSK group of companies at the time of the experimental work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Giannelli, C., Necchi, F., Palmieri, E. et al. Quality by Design Framework Applied to GMMA Purification. AAPS J 26, 32 (2024). https://doi.org/10.1208/s12248-024-00902-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-024-00902-0