Abstract
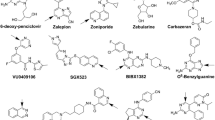
CYP1A1 is a cytochrome P450 family 1 enzyme that is mostly expressed in the extrahepatic tissues. To understand the CYP1A1 contribution to drug clearance in humans, we examined the in vitro–in vivo extrapolation (IVIVE) of intrinsic clearance (CLint) for a set of drugs that are in vitro CYP1A1 substrates. Despite being strong in vitro CYP1A1 substrates, 82% of drugs gave good IVIVE with predicted CLint within 2–3-fold of the observed values using human liver microsomes and hepatocytes, suggesting they were not in vivo CYP1A1 substrates due to the lack of extrahepatic contribution to CLint. Only three drugs (riluzole, melatonin and ramelteon) that are CYP1A2 substrates yielded significant underprediction of in vivo CLint up to 11-fold. The fold of CLint underprediction was linearly proportional to human recombinant CYP1A1 (rCYP1A1) CLint, indicating they were likely to be in vivo CYP1A1 substrates. Using these three substrates, a calibration curve can be developed to enable direct translation from in vitro rCYP1A1 CLint to in vivo extrahepatic contributions in humans. In vivo CYP1A1 substrates are planar and small, which is consistent with the structure of the active site. This is in contrast to the in vitro substrates, which include large and nonplanar molecules, suggesting rCYP1A1 is more accessible than what is in vivo. The impact of CYP1A1 on first-pass intestinal metabolism was also evaluated and shown to be minimal. This is the first study providing new insights on in vivo translation of CYP1A1 contributions to human clearance using in vitro rCYP1A1 data.
Graphical Abstract




Similar content being viewed by others
References
Sridhar J, Goyal N, Liu J, Foroozesh M. Review of ligand specificity factors for CYP1A subfamily enzymes from molecular modeling studies reported to-date. Molecules (Basel, Switzerland). 2017;22(7). https://doi.org/10.3390/molecules22071143.
Shimada T, Yun CH, Yamazaki H, Gautier JC, Beaune PH, Guengerich FP. Characterization of human lung microsomal cytochrome P-450 1A1 and its role in the oxidation of chemical carcinogens. Mol Pharmacol. 1992;41(5):856–64.
Klomp F, Wenzel C, Drozdzik M, Oswald S. Drug-drug interactions involving intestinal and hepatic CYP1A enzymes. Pharmaceutics. 2020;12(12):1201. https://doi.org/10.3390/pharmaceutics12121201.
Yengi LG, Xiang Q, Pan J, Scatina J, Kao J, Ball SE, et al. Quantitation of cytochrome P450 mRNA levels in human skin. Anal Biochem. 2003;316(1):103–10. https://doi.org/10.1016/s0003-2697(03)00042-3.
van de Kerkhof EG, Graaf IAMd, Ungell A-LB, Groothuis GMM. Induction of metabolism and transport in human intestine: validation of precision-cut slices as a tool to study induction of drug metabolism in human intestine in vitro. Drug Metab Dispos. 2008;36(3):604–13. https://doi.org/10.1124/dmd.107.018820.
Nishimura M, Yaguti H, Yoshitsugu H, Naito S, Satoh T. Tissue distribution of mRNA expression of human cytochrome P450 isoforms assessed by high-sensitivity real-time reverse transcription PCR. Yakugaku Zasshi. 2003;123(5):369–75. https://doi.org/10.1248/yakushi.123.369.
Hakkola J, Raunio H, Purkunen R, Pelkonen O, Saarikoski S, Cresteil T, et al. Detection of cytochrome P450 gene expression in human placenta in first trimester of pregnancy. Biochem Pharmacol. 1996;52(2):379–83. https://doi.org/10.1016/0006-2952(96)00216-x.
Walsh AA, Szklarz GD, Scott EE. Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism. J Biol Chem. 2013;288(18):12932–43. https://doi.org/10.1074/jbc.M113.452953.
Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270(1):414–23.
Smith CAD, Smith G, Wolf CR. Genetic polymorphisms in xenobiotic metabolism. Eur J Cancer. 1994;30(13):1921–35. https://doi.org/10.1016/0959-8049(94)00382-F.
Stiborová M, Martínek V, Rýdlová H, Koblas T, Hodek P. Expression of cytochrome P450 1A1 and its contribution to oxidation of a potential human carcinogen 1-phenylazo-2-naphthol (Sudan I) in human livers. Cancer Lett. 2005;220(2):145–54. https://doi.org/10.1016/j.canlet.2004.07.036.
Wegler C, Wiśniewski JR, Robertsen I, Christensen H, KristofferHertel J, Hjelmesaeth J, et al. Drug disposition protein quantification in matched human jejunum and liver from donors with obesity. Clin Pharmacol Ther. 2022;111(5):1142–54. https://doi.org/10.1002/cpt.2558.
Wahid M, Mahjabeen I, Baig RM, Kayani MA. Expression of CYP1A1 and GSTP1 in human brain tumor tissues in Pakistan. Asian Pac J Cancer Prev: APJCP. 2013;14(12):7187–91. https://doi.org/10.7314/apjcp.2013.14.12.7187.
Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC. The human intestinal cytochrome P450 “pie.” Drug Metab Dispos. 2006;34(5):880–6. https://doi.org/10.1124/dmd.105.008672.
Ma X, Idle JR, Krausz KW, Gonzalez FJ. Metabolism of melatonin by human cytochromes p450. Drug Metab Dispos. 2005;33(4):489–94. https://doi.org/10.1124/dmd.104.002410.
Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT. Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology. 2003;144(8):3382–98. https://doi.org/10.1210/en.2003-0192.
Becker C, Frey R, Unger S, Thomas D, Reber M, Weimann G, et al. Pharmacokinetic interaction of riociguat with ketoconazole, clarithromycin, and midazolam. Pulm Circ. 2016;6(Suppl 1):S49-57. https://doi.org/10.1086/685016.
Lang D, Radtke M, Bairlein M. Highly Variable expression of CYP1A1 in human liver and impact on pharmacokinetics of riociguat and granisetron in humans. Chem Res Toxicol. 2019;32(6):1115–22. https://doi.org/10.1021/acs.chemrestox.8b00413.
Werlinder V, Backlund M, Zhukov A, Ingelman-Sundberg M. Transcriptional and post-translational regulation of CYP1A1 by primaquine. J Pharmacol Exp Ther. 2001;297(1):206–14.
Santes-Palacios R, Ornelas-Ayala D, Cabanas N, Marroquin-Perez A, Hernandez-Magana A, Olguin-Reyes SdR, et al. Regulation of human cytochrome P4501A1 (hCYP1A1): a plausible target for chemoprevention? BioMed Res Int. 2016;5341081/1. https://doi.org/10.1155/2016/5341081.
Lekas P, Tin KL, Lee C, Prokipcak RD. The human cytochrome P450 1A1 mRNA is rapidly degraded in HepG2 cells. Arch Biochem Biophys. 2000;384(2):311–8. https://doi.org/10.1006/abbi.2000.2115.
Zhao X, Wang Z, Wang Y, Zhang H, Blode H, Yoshikawa K, et al. Pharmacokinetics of the soluble guanylate cyclase stimulator riociguat in healthy young Chinese male non-smokers and smokers: results of a randomized, double-blind, placebo-controlled study. Clin Pharmacokinet. 2016;55(5):615–24. https://doi.org/10.1007/s40262-015-0337-4.
Dong D, Wu B, Chow D, Hu M. Substrate selectivity of drug-metabolizing cytochrome P450s predicted from crystal structures and in silico modeling. Drug Metab Rev. 2012;44(2):192–208. https://doi.org/10.3109/03602532.2011.645580.
Bart AG, Takahashi RH, Wang X, Scott EE. Human cytochrome P450 1A1 adapts active site for atypical nonplanar substrate. Drug Metab Dispos. 2020;48(2):86–92. https://doi.org/10.1124/dmd.119.089607.
Saravanakumar A, Sadighi A, Ryu R, Akhlaghi F. Physicochemical properties, biotransformation, and transport pathways of established and newly approved medications: a systematic review of the top 200 most prescribed drugs vs. the FDA-approved drugs between 2005 and 2016. Clin Pharmacokinet. 2019;58(10):1281–94. https://doi.org/10.1007/s40262-019-00750-8.
Lang D, Jungmann N. Drug library screening for human CYP1A1 substrates. Drug Metab Pharmacokinet. 2020;35(1, Supplement):S28. https://doi.org/10.1016/j.dmpk.2020.04.022.
Takahashi RH, Wang X, Segraves NL, Wang J, Chang JH, Khojasteh SC, et al. CYP1A1-mediated intramolecular rearrangement of aminoazepane in GDC-0339. Drug Metab Dispos. 2017;45(10):1084–92. https://doi.org/10.1124/dmd.117.076786.
Tess D, Chang GC, Keefer C, Carlo A, Jones R, Di L. In vitro-in vivo extrapolation and scaling factors for clearance of human and preclinical species with liver microsomes and hepatocytes. AAPS J. 2023;25(3):40. https://doi.org/10.1208/s12248-023-00800-x.
Certara Drug Interaction Solution https://www.druginteractionsolutions.org/
Andersen LP, Werner MU, Rosenkilde MM, Harpsøe NG, Fuglsang H, Rosenberg J, et al. Pharmacokinetics of oral and intravenous melatonin in healthy volunteers. BMC Pharmacol Toxicol. 2016;17:8. https://doi.org/10.1186/s40360-016-0052-2.
Price E, Kalvass JC, DeGoey D, Hosmane B, Doktor S, Desino K. Global analysis of models for predicting human absorption: QSAR, in vitro, and preclinical models. J Med Chem. 2021;64(13):9389–403. https://doi.org/10.1021/acs.jmedchem.1c00669.
Mebendazole HB. In: Enna SJ, Bylund DB, editors. xPharm: the comprehensive pharmacology reference. New York: Elsevier; 2007. p. 1–5.
Rosenkranz B, Winkelmann BR, Parnham MJ. Clinical pharmacokinetics of molsidomine. Clin Pharmacokinet. 1996;30(5):372–84. https://doi.org/10.2165/00003088-199630050-00004.
Keefer C, Chang G, Carlo A, Novak JJ, Banker M, Carey J, et al. Mechanistic insights on clearance and inhibition discordance between liver microsomes and hepatocytes when clearance in liver microsomes is higher than in hepatocytes. Eur J Pharm Sci. 2020;155: 105541. https://doi.org/10.1016/j.ejps.2020.105541.
Xu B, Ding J, Chen KX, Miao ZH, Huang H, Liu H, et al. Advances in cancer chemotherapeutic drug research in China. Recent Adv Cancer Res Ther. 2012;287-350. https://doi.org/10.1016/b978-0-12-397833-2.00012-1.
Davies M, Peramuhendige P, King L, Golding M, Kotian A, Penney M, et al. Evaluation of in vitro models for assessment of human intestinal metabolism in drug discovery. Drug Metab Dispos. 2020;DMD-AR-2020-000111. https://doi.org/10.1124/dmd.120.000111.
Zhang Z, Li Y, Stearns RA, Ortiz De Montellano PR, Baillie TA, Tang W. Cytochrome P450 3A4-mediated oxidative conversion of a cyano to an amide group in the metabolism of pinacidil. Biochemistry. 2002;41(8):2712–8. https://doi.org/10.1021/bi0119971.
Sarkar M, Grossman RG, Toups EG, Chow DS. Rational design and development of a stable liquid formulation of riluzole and its pharmacokinetic evaluation after oral and IV administrations in rats. Eur J Pharm Sci. 2018;125:1–10. https://doi.org/10.1016/j.ejps.2018.09.004.
Ferreira MA Jr, Azevedo H, Mascarello A, Segretti ND, Russo E, Russo V, et al. Discovery of ACH-000143: a novel potent and peripherally preferred melatonin receptor agonist that reduces liver triglycerides and steatosis in diet-induced obese rats. J Med Chem. 2021;64(4):1904–29. https://doi.org/10.1021/acs.jmedchem.0c00627.
Tess DA, Ryu S, Di L. In vitro - in vivo extrapolation of hepatic clearance in preclinical species. Pharm Res. 2022;39(7):1615–32. https://doi.org/10.1007/s11095-022-03205-1.
Nakamura H, Ariyoshi N, Okada K, Nakasa H, Nakazawa K, Kitada M. CYP1A1 is a major enzyme responsible for the metabolism of granisetron in human liver microsomes. Curr Drug Metab. 2005;6(5):469–80. https://doi.org/10.2174/138920005774330666.
McGinnity DF, Soars MG, Urbanowicz RA, Riley RJ. Evaluation of fresh and cryopreserved hepatocytes as in vitro drug metabolism tools for the prediction of metabolic clearance. Drug Metab Dispos. 2004;32(11):1247–53. https://doi.org/10.1124/dmd.104.000026.
Sohlenius-Sternbeck AK, Afzelius L, Prusis P, Neelissen J, Hoogstraate J, Johansson J, et al. Evaluation of the human prediction of clearance from hepatocyte and microsome intrinsic clearance for 52 drug compounds. Xenobiotica. 2010;40(9):637–49. https://doi.org/10.3109/00498254.2010.500407.
Corrigan BW, Nicholls B, Thakrar B, Lam R, Grosse C, Alianti J, et al. Heterogeneity in systemic availability of ondansetron and granisetron following oral administration. Drug Metab Dispos. 1999;27(1):110–2.
Acknowledgements
The author greatly appreciates the help of Sophia M. Shi in editing the manuscript and Joy Yang in providing property calculation and insights, as well as many Pfizer and external colleagues for their helpful discussion of the topic.
Funding
This study was sponsored by Pfizer Inc., New York, NY, USA.
Author information
Authors and Affiliations
Contributions
L.D. designed the study, analyzed the data, and wrote the paper.
Corresponding author
Ethics declarations
Conflict of Interest
Li Di is an employee of Pfizer Inc., New York, NY, USA, and holds stock or stock options.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Di, L. Quantitative Translation of Substrate Intrinsic Clearance from Recombinant CYP1A1 to Humans. AAPS J 25, 98 (2023). https://doi.org/10.1208/s12248-023-00863-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-023-00863-w